Abstract
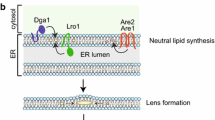
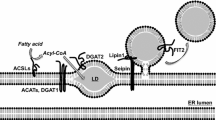
The lipid droplet (LD) is a highly dynamic organelle that maintains cellular lipid homeostasis in addition to storing energy sources. Current research suggests LDs are responsible for the transportation, storage and lipolysis-driven mobilization of lipids within cells. Here, we review the landscape of evidence for LD involvement in regulating lipid homeostasis. LD interactions with other organelles, particularly the endoplasmic reticulum, mitochondria, lysosomes (or vacuoles in yeast), and peroxisomes, highlight their importance for lipid transfer and metabolism.

Similar content being viewed by others
References
Farese, R.V., Jr. and Walther, T.C., Lipid droplets finally get a little R-E-S-P-E-C-T, Cell, 2009, vol. 139, pp. 855–860. doi:10.1016/j.cell.2009.11.005
Murphy, D.J., The biogenesis and functions of lipid bodies in animals, plants and microorganisms, Progress in Lipid Research, 2001, vol. 40, pp. 325–438. doi: 10.1016/S0163-7827(01)00013-3
Kalscheuer, R., Wältermann, M., Alvarez, M., and Steinbüchel, A., Preparative isolation of lipid inclusions from Rhodococcus opacus and Rhodococcus ruber and identification of granule-associated proteins, Arch. Microbiol., 2001 vol. 177, pp. 20–28. doi: 10.1007/s00203-001-0355-5
Moellering, E. and Benning, C., RNA interference silencing of a major lipid droplet protein affects lipid droplet size in Chlamydomonas reinhardtii, Eukaryot Cell, 2010, vol. 9, pp. 97–106. doi: 10.1128/EC.00203-09
Athenstaedt, K., Zweytick, D., Jandrositz, A., Kohlwein, S., et al., Identification and characterization of major lipid particle proteins of the yeast Saccharomyces cerevisiae, J. Bacteriol., 1999, vol. 181, pp. 6441–6448. doi: 10.1128/JB.181.20.6441-6448.1999
Grillitsch, K., Connerth, M., Köfeler, H., Arrey, T., et al., Lipid particles/droplets of the yeast Saccharomyces cerevisiae revisited: lipidome meets proteome, Biochim. Biophys. Acta, 2011, vol. 1811, pp. 1165–1176. doi: 10.1016/j.bbalip.2011.07.015
Zhang, P., Na, H., Liu, Z., Zhang, S., et al., Proteomic study and marker protein identification of Caenorhabditis elegans lipid droplets, Mol. Cell. Proteomics, 2012, vol. 11(8), pp. 317–328. doi: 10.1074/mcp.M111.016345
Beller, M., Riedel, D., Jänsch, L., Dieterich, G., et al., Characterization of the Drosophila lipid droplet subproteome, Mol. Cell. Proteomics, 2006, vol. 5, pp. 1082–1094. doi: 10.1074/mcp.M600011-MCP200
Cermelli, S., Guo, Y., Gross, S., and Welte, M., The lipid-droplet proteome reveals that droplets are a protein-storage depot, Curr. Biol., 2006, vol. 16, pp. 1783–1795. doi: 10.1016/j.cub.2006.07.062
Jolivet, P., Roux, E., D’Andrea, S., Davanture, M., et al., Protein composition of oil bodies in Arabidopsis thaliana ecotype WS, Plant Physiol. Biochem., 2004, vol. 42, pp. 501–509. doi: 10.1016/j.plaphy.2004.04.006
Lin, L., Liao, P., Yang, H., and Tzen, J., Determination and analyses of the N-termini of oil-body proteins, steroleosin, caleosin and oleosin, Plant Physiol. Biochem., 2005, vol. 43, pp. 770–776. doi: 10.1016/j.plaphy.2005.07.008
Katavic, V., Agrawal, G., Hajduch, M., Harris, S., et al., Protein and lipid composition analysis of oil bodies from two Brassica napus cultivars, Proteomics, 2006, vol. 6, pp. 4586–4598. doi: 10.1002/pmic.200600020
Tian, J., Zhang, J., Yu, E., Sun, J., et al., Identification and analysis of lipid droplet-related proteome in the adipose tissue of grass carp (Ctenopharyngodon idella) under fed and starved conditions, Comp. Biochem. Physiol. Part D Genomics Proteomics, 2020, vol. 36, pp. 100710. doi: 10.1016/j.cbd.2020.100710
Zhang, H., Wang, Y., Li, Y., Yu, J., et al., Proteome of skeletal muscle lipid droplet reveals association with mitochondria and apolipoprotein a-I, J. Proteome Res., 2011, vol. 10, pp. 4757–4768. doi: 10.1021/pr200553c
Yu, J., Zhang, L., Li, Y., Zhu, X., et al., The adrenal lipid droplet is a new site for steroid hormone metabolism, Proteomics, 2018, vol. 18, p. e1800136. doi: 10.1002/pmic.201800136
Wan, H., Melo, R., Jin, Z., Dvorak, A., et al., Roles and origins of leukocyte lipid bodies: proteomic and ultrastructural studies, FASEB J, 2007, vol. 21, pp. 167–178. doi: 10.1096/fj.06-6711com
Fujimoto, Y., Itabe, H., Sakai, J., Makita, M., et al., Identification of major proteins in the lipid droplet-enriched fraction isolated from the human hepatocyte cell line HuH7, Biochim. Biophys. Acta, 2004, vol. 1644, pp. 47–59. doi: 10.1016/j.bbamcr.2003.10.018
Murphy, S., Martin, S., and Parton, R.G., Lipid droplet-organelle interactions; sharing the fats, Biochim. Biophys. Acta, 2009, vol. 1791, pp. 441–447. doi: 10.1016/j.bbalip.2008.07.004
Zehmer, J.K., Huang, Y., Peng, G., Pu, J., et al., A role for lipid droplets in inter-membrane lipid traffic, Proteomics, 2009, vol. 9, pp. 914–921. doi: 10.1002/pmic.200800584
Barbosa, A.D., Savage, D.B., and Siniossoglou, S., Lipid droplet-organelle interactions: emerging roles in lipid metabolism, Curr. Opin. Cell. Biol., 2015, vol. 35, pp. 91–97. doi: 10.1016/j.ceb.2015.04.017
Walther, T.C. and Farese, R.V., Jr., Lipid droplets and cellular lipid metabolism, Annu Rev. Biochem., 2012, vol. 81, pp. 687–714. doi: 10.1146/annurev-biochem-061009-102430
Blanchette-Mackie, E.J., Dwyer, N.K., Barber, T., Coxey, R.A., et al., Perilipin is located on the surface layer of intracellular lipid droplets in adipocytes, J. Lipid Res., 1995, vol. 36, pp. 1211–1226.
Robenek, H., Robenek, M.J., and Troyer, D., PAT family proteins pervade lipid droplet cores, J. Lipid Res., 2005, vol. 46, pp. 1331–1338. doi: 10.1194/jlr.M400323-JLR200
Wilfling, F., Wang, H., Haas, J.T., Krahmer, N., et al., Triacylglycerol synthesis enzymes mediate lipid droplet growth by relocalizing from the ER to lipid droplets, Dev. Cell., 2013, vol. 24, pp. 384–399. doi: 10.1016/j.devcel.2013.01.013
Waltermann, M., Hinz, A., Robenek, H., Troyer, D., et al., Mechanism of lipid-body formation in prokaryotes: how bacteria fatten up, Mol. Microbiol., 2005, vol. 55, pp. 750–763. doi: 10.1111/j.1365-2958.2004.04441.x
Choudhary, V., Ojha, N., Golden, A., and Prinz, W.A., A conserved family of proteins facilitates nascent lipid droplet budding from the ER, J. Cell. Biol., 2015, vol. 211, pp. 261–271. doi: 10.1083/jcb.201505067
Walther, T.C., Chung, J., and Farese, R.V., Jr., Lipid Droplet Biogenesis, Annu Rev. Cell. Dev Biol, 2017, vol. 33, pp. 491–510. doi: 10.1146/annurev-cellbio-100616-060608
Qi, Y., Sun, L., and Yang, H., Lipid droplet growth and adipocyte development: mechanistically distinct processes connected by phospholipids, Biochim. Biophys. Acta Mol. Cell. Biol. Lipids, 2017, vol. 1862, pp. 1273–1283. doi: 10.1016/j.bbalip.2017.06.016
Jacquier, N., Choudhary, V., Mari, M., Toulmay, A., et al., Lipid droplets are functionally connected to the endoplasmic reticulum in Saccharomyces cerevisiae, J. Cell. Sci., 2011, vol. 124, pp. 2424–2437. doi: 10.1242/jcs.076836
Kassan, A., Herms, A., Fernandez-Vidal, A., Bosch, M., et al., Acyl-CoA synthetase 3 promotes lipid droplet biogenesis in ER microdomains, J. Cell. Biol., 2013, vol. 203, pp. 985–1001. doi: 10.1083/jcb.201305142
Joshi, A.S., Zhang, H., and Prinz, W.A., Organelle biogenesis in the endoplasmic reticulum, Nat. Cell. Biol., 2017, vol. 19, pp. 876–882. doi: 10.1038/ncb3579
Thiam, A.R., Antonny, B., Wang, J., Delacotte, J., et al., COPI buds 60-nm lipid droplets from reconstituted water–phospholipid–triacylglyceride interfaces, suggesting a tension clamp function, Proc. Natl. Acad. Sci. USA, 2013, vol. 110, pp. 13244–13249. doi: 10.1073/pnas.1307685110
Wilfling, F., Thiam, A.R., Olarte, M.J., Wang, J., et al., Arf1/COPI machinery acts directly on lipid droplets and enables their connection to the ER for protein targeting, Elife, 2014, vol. 3, pp. e01607. doi: 10.7554/eLife.01607
Magre, J., Delepine, M., Khallouf, E., Gedde-Dahl, T., Jr., et al., Identification of the gene altered in Berardinelli–Seip congenital lipodystrophy on chromosome 11q13, Nat. Genet., 2001, vol. 28, pp. 365–370. doi: 10.1038/ng585
Wang, H., Becuwe, M., Housden, B.E., Chitraju, C., et al., Seipin is required for converting nascent to mature lipid droplets, Elife, 2016, vol. 5, p. e16582. doi: 10.7554/eLife.16582
Grippa, A., Buxo, L., Mora, G., Funaya, C., et al., The seipin complex Fld1/Ldb16 stabilizes ER–lipid droplet contact sites, J. Cell. Biol., 2015, vol. 211, pp. 829–844. doi:10.1083/jcb.201502070
Pagac, M., Cooper, D.E., Qi, Y., Lukmantara, I.E., et al., SEIPIN regulates lipid droplet expansion and adipocyte development by modulating the activity of glycerol-3-phosphate acyltransferase, Cell. Rep., 2016, vol. 17, pp. 1546–1559. doi:10.1016/j.celrep.2016.10.037
Henne, W.M., Reese, M.L., and Goodman, J.M., The assembly of lipid droplets and their roles in challenged cells, Embo j., 2019, vol. 38, p. e98947. doi:10.15252/embj.2019101816
Salo, V.T., Li, S., Vihinen, H., Holtta-Vuori, M., et al., Seipin facilitates triglyceride flow to lipid droplet and counteracts droplet ripening via endoplasmic reticulum contact, Dev. Cell., 2019, vol. 50, pp. 478–493.e479. doi: 10.1016/j.devcel.2019.05.016
Xu, D., Li, Y., Wu, L., Li, Y., et al., Rab18 promotes lipid droplet (LD) growth by tethering the ER to LDs through SNARE and NRZ interactions, J. Cell. Biol., 2018, vol. 217, pp. 975–995. doi: 10.1083/jcb.201704184
Li, D., Zhao, Y.G., Li, D., Zhao, H., et al., The ER-localized protein DFCP1 modulates ER–lipid droplet contact formation, Cell. Rep., 2019, vol. 27, pp. 343–358.e345. doi: 10.1016/j.celrep.2019.03.025
Wilfling, F., Haas, J.T., Walther, T.C., and Farese, R.V., Jr., Lipid droplet biogenesis, Curr. Opin. Cell. Biol., 2014, vol. 29, pp. 39–45. doi: 10.1016/j.ceb.2014.03.008
Yu, J. and Li, P., The size matters: regulation of lipid storage by lipid droplet dynamics, Sci. China Life. Sci., 2017, vol. 60, pp. 46–56. doi: 10.1007/s11427-016-0322-x
Murphy, S., Martin, S., and Parton, R.G., Quantitative analysis of lipid droplet fusion: inefficient steady state fusion but rapid stimulation by chemical fusogens, PLoS One, 2010, vol. 5, p. e15030. doi: 10.1371/journal.pone.0015030
Fei, W., Shui, G., Zhang, Y., Krahmer, N., et al., A role for phosphatidic acid in the formation of “supersized” lipid droplets, PLoS Genet., 2011, vol. 7, pp. e1002201. doi: 10.1371/journal.pgen.1002201
Guo, Y., Walther, T.C., Rao, M., Stuurman, N., et al., Functional genomic screen reveals genes involved in lipid-droplet formation and utilization, Nature, 2008, vol. 453, pp. 657–661. doi: 10.1038/nature06928
Xu, W., Wu, L., Yu, M., Chen, F.J., et al., Differential roles of cell death-inducing DNA fragmentation factor-alpha-like effector (CIDE) proteins in promoting lipid droplet fusion and growth in subpopulations of hepatocytes, J. Biol. Chem., 2016, vol. 291, pp. 4282–4293. doi:10.1074/jbc.M115.701094
Gong, J., Sun, Z., Wu, L., Xu, W., et al., Fsp27 promotes lipid droplet growth by lipid exchange and transfer at lipid droplet contact sites, J. Cell. Biol., 2011, vol. 195, pp. 953–963. doi:10.1083/jcb.201104142
Jambunathan, S., Yin, J., Khan, W., Tamori, Y., et al., FSP27 promotes lipid droplet clustering and then fusion to regulate triglyceride accumulation, PLoS One, 2011, vol. 6, pp. e28614. doi:10.1371/journal.pone.0028614
Bostrom, P., Andersson, L., Rutberg, M., Perman, J., et al., SNARE proteins mediate fusion between cytosolic lipid droplets and are implicated in insulin sensitivity, Nat. Cell. Biol., 2007, vol. 9, pp. 1286–1293. doi:10.1038/ncb1648
Rambold, A.S., Cohen, S., and Lippincott-Schwartz, J., Fatty acid trafficking in starved cells: regulation by lipid droplet lipolysis, autophagy, and mitochondrial fusion dynamics, Dev. Cell., 2015, vol. 32, pp. 678–692. doi:10.1016/j.devcel.2015.01.029
Axe, E.L., Walker, S.A., Manifava, M., Chandra, P., et al., Autophagosome formation from membrane compartments enriched in phosphatidylinositol 3-phosphate and dynamically connected to the endoplasmic reticulum, J. Cell. Biol., 2008, vol. 182, pp. 685–701. doi:10.1083/jcb.200803137
Hayashi-Nishino, M., Fujita, N., Noda, T., Yamaguchi, A., et al., A subdomain of the endoplasmic reticulum forms a cradle for autophagosome formation, Nat. Cell. Biol., 2009, vol. 11, pp. 1433–1437. doi:10.1038/ncb1991
Kristensen, A.R., Schandorff, S., Hoyer-Hansen, M., Nielsen, M.O., et al., Ordered organelle degradation during starvation-induced autophagy, Mol. Cell. Proteomics, 2008, vol. 7, pp. 2419–2428. doi:10.1074/mcp.M800184-MCP200
Singh, R., Kaushik, S., Wang, Y., Xiang, Y., et al., Autophagy regulates lipid metabolism, Nature, 2009, vol. 458, pp. 1131–1135. doi:10.1038/nature07976
Yla-Anttila, P., Vihinen, H., Jokitalo, E., and Eskelinen, E.L., 3D tomography reveals connections between the phagophore and endoplasmic reticulum, Autophagy, 2009, vol. 5, pp. 1180–1185. doi:10.4161/auto.5.8.10274
Lass, A., Zimmermann, R., Oberer, M., and Zechner, R., Lipolysis—a highly regulated multi-enzyme complex mediates the catabolism of cellular fat stores, Prog. Lipid Res., 2011, vol. 50, pp. 14–27. doi:10.1016/j.plipres.2010.10.004
Wang, S., Soni, K.G., Semache, M., Casavant, S., et al., Lipolysis and the integrated physiology of lipid energy metabolism, Mol. Genet. Metab., 2008, vol. 95, pp. 117–126. doi:10.1016/j.ymgme.2008.06.012
Zechner, R., Zimmermann, R., Eichmann, T.O., Kohlwein, S.D., et al., FAT SIGNALS—lipases and lipolysis in lipid metabolism and signaling, Cell. Metab., 2012, vol. 15, pp. 279–291. doi:10.1016/j.cmet.2011.12.018
Brasaemle, D.L., Rubin, B., Harten, I.A., Gruia-Gray, J., et al., Perilipin A increases triacylglycerol storage by decreasing the rate of triacylglycerol hydrolysis, J. Biol. Chem., 2000, vol. 275, pp. 38486–38493. doi:10.1074/jbc.M007322200
Londos, C., Brasaemle, D.L., Schultz, C.J., Adler-Wailes, D.C., et al., On the control of lipolysis in adipocytes, Ann. NY Acad. Sci., 1999, vol. 892, pp. 155–168. doi:10.1111/j.1749-6632.1999.tb07794.x
Granneman, J.G., Moore, H.P., Mottillo, E.P., Zhu, Z., et al., Interactions of perilipin-5 (Plin5) with adipose triglyceride lipase, J. Biol. Chem., 2011, vol. 286, pp. 5126–5135. doi:10.1074/jbc.M110.180711
Weidberg, H., Shvets, E., and Elazar, Z., Biogenesis and cargo selectivity of autophagosomes, Annu. Rev. Biochem., 2011, vol. 80, pp. 125–156. doi:10.1146/annurev-biochem-052709-094552
Zechner, R., Madeo, F., and Kratky, D., Cytosolic lipolysis and lipophagy: two sides of the same coin, Nat. Rev. Mol. Cell. Biol., 2017, vol. 18, pp. 671–684. doi:10.1038/nrm.2017.76
Valm, A.M., Cohen, S., Legant, W.R., Melunis, J., et al., Applying systems-level spectral imaging and analysis to reveal the organelle interactome, Nature, 2017, vol. 546, pp. 162–167. doi:10.1038/nature22369
Kaushik, S. and Cuervo, A.M., Degradation of lipid droplet-associated proteins by chaperone-mediated autophagy facilitates lipolysis, Nat. Cell. Biol., 2015, vol. 17, pp. 759–770. doi:10.1038/ncb3166
Schroeder, B., Schulze, R.J., Weller, S.G., Sletten, A.C., et al., The small GTPase Rab7 as a central regulator of hepatocellular lipophagy, Hepatology, 2015, vol. 61, pp. 1896–1907. doi:10.1002/hep.27667
Nguyen, T.B., Louie, S.M., Daniele, J.R., Tran, Q., et al., DGAT1-dependent lipid droplet biogenesis protects mitochondrial function during starvation-induced autophagy, Dev. Cell., 2017, vol. 42, pp. 9–21. e25. doi:10.1016/j.devcel.2017.06.003
Barbosa, A.D. and Siniossoglou, S., Spatial distribution of lipid droplets during starvation: Implications for lipophagy, Commun. Integr. Biol., 2016, vol. 9, p. e1183854. doi:10.1080/19420889.2016.1183854
Hariri, H., Rogers, S., Ugrankar, R., Liu, Y.L., et al., Lipid droplet biogenesis is spatially coordinated at ER-vacuole contacts under nutritional stress, EMBO Rep., 2018, vol. 19, pp. 57–72. doi:10.15252/embr.201744815
Finn, P.F. and Dice, J.F., Proteolytic and lipolytic responses to starvation, Nutrition, 2006, vol. 22, pp. 830–844. doi:10.1016/j.nut.2006.04.008
Herms, A., Bosch, M., Reddy, B.J., Schieber, N.L., et al., AMPK activation promotes lipid droplet dispersion on detyrosinated microtubules to increase mitochondrial fatty acid oxidation, Nat. Commun., 2015, vol. 6, pp. 7176. doi:10.1038/ncomms8176
Pu, J., Ha, C.W., Zhang, S., Jung, J.P., et al., Interactomic study on interaction between lipid droplets and mitochondria, Protein Cell, 2011, vol. 2, pp. 487–496. doi:10.1007/s13238-011-1061-y
Benador, I.Y., Veliova, M., Mahdaviani, K., Petcherski, A., et al., Mitochondria bound to lipid droplets have unique bioenergetics, composition, and dynamics that support lipid droplet expansion, Cell Metab., 2018, vol. 27, pp. 869–885. e6. doi:10.1016/j.cmet.2018.03.003
Wang, H., Sreenivasan, U., Hu, H., Saladino, A., et al., Perilipin 5, a lipid droplet associated protein, provides physical and metabolic linkage to mitochondria, J. Lipid Res., 2011, vol. 52, pp. 2159–2168. doi:10.1194/jlr.M017939
Jagerstrom, S., Polesie, S., Wickstrom, Y., Johansson, B.R., et al., Lipid droplets interact with mitochondria using SNAP23, Cell. Biol. Int., 2009, vol. 33, pp. 934–940. doi:10.1016/j.cellbi.2009.06.011
Dirkx, R., Vanhorebeek, I., Martens, K., Schad, A., et al., Absence of peroxisomes in mouse hepatocytes causes mitochondrial and ER abnormalities, Hepatology, 2005, vol. 41, pp. 868–878. doi:10.1002/hep.20628
Thazar-Poulot, N., Miquel, M., Fobis-Loisy, I., and Gaude, T., Peroxisome extensions deliver the Arabidopsis SDP1 lipase to oil bodies, Proc. Natl. Acad. Sci. USA, 2015, vol. 112, pp. 4158–4163. doi:10.1073/pnas.1403322112
Binns, D., Januszewski, T., Chen, Y., Hill, J., et al., An intimate collaboration between peroxisomes and lipid bodies, J. Cell. Biol., 2006, vol. 173, pp. 719–731. doi:10.1083/jcb.200511125
Kory, N., Farese, R.V., Jr., and Walther, T.C., Targeting Fat: Mechanisms of protein localization to lipid droplets, Trends Cell. Biol., 2016, vol. 26, pp. 535–546. doi:10.1016/j.tcb.2016.02.007
Hynynen, R., Suchanek, M., Spandl, J., Back, N., et al., OSBP-related protein 2 is a sterol receptor on lipid droplets that regulates the metabolism of neutral lipids, J. Lipid Res., 2009, vol. 50, pp. 1305–1315. doi:10.1194/jlr.M800661-JLR200
Ohsaki, Y., Kawai, T., Yoshikawa, Y., Cheng, J., et al., PML isoform II plays a critical role in nuclear lipid droplet formation, J. Cell. Biol., 2016, vol. 212, pp. 29–38. doi:10.1083/jcb.201507122
Romanauska, A. and Kohler, A., The inner nuclear membrane is a metabolically active territory that generates nuclear lipid droplets, Cell, 2018, vol. 174, pp. 700–715. e718. doi:10.1016/j.cell.2018.05.047
Farese, R.V., Jr. and Walther, T.C., Lipid droplets go nuclear, J. Cell. Biol., 2016, vol. 212, pp. 7–8. doi:10.1083/jcb.201512056
Welte, M.A., Expanding roles for lipid droplets, Curr. Biol., 2015, vol. 25, pp. R470–481. doi:10.1016/j.cub.2015.04.004
Funding
This work was supported by the Natural Science Foundation of Guangdong Province, China (Grant Number 202015150224).
Author information
Authors and Affiliations
Corresponding authors
Rights and permissions
About this article
Cite this article
Huang, J., Chen, X., Zhang, F. et al. Lipid Droplet Metabolism Across Eukaryotes: Evidence from Yeast to Humans. J Evol Biochem Phys 56, 396–405 (2020). https://doi.org/10.1134/S0022093020050026
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1134/S0022093020050026