Abstract
The Silver–Russell syndrome (SRS) is a rare disorder characterized by heterogeneous clinical features, including growth retardation, typical facial dysmorphisms, and body asymmetry. Genetic alterations causative of SRS mostly affect imprinted genes located on chromosomes 7 or 11. Hypomethylation of the Imprinting Center 1 (IC1) of the chromosome 11p15.5 is the most common cause of SRS, while the Imprinting Center 2 (IC2) has been more rarely involved. Specifically, maternally inherited 11p15.5 deletions including the IC2 have been associated with the Beckwith–Wiedemann Syndrome (BWS), while paternal deletions with a variable spectrum of phenotypes. Here, we describe the case of a girl with a mild SRS phenotype associated with a paternally inherited 1.4 kb deletion of IC2. The father of the proband inherited the deletion from his mother and showed normal growth, while the paternal grandmother had the deletion on her paternal chromosome and exhibited short stature. Together with previous findings obtained in mouse and humans, our data support the notion that deletion of the paternal copy of IC2 can cause SRS.
Similar content being viewed by others
Introduction
Approximately 1% of the human genes is imprinted, i.e. being characterized by differential epigenetic modification causing diverse expression of the maternally-derived and the paternally-derived alleles [1]. Imprinted genes are largely organized in conserved clusters that are fundamental for normal growth and development; perturbation of imprinted gene dosage caused by genetic or epigenetic alterations results in a heterogenous class of diseases, better known as imprinting disorders. The latter are mostly associated with developmental abnormalities, fetal growth, and metabolic alterations as well as specific neurological behaviors [2]. Imprinted gene clusters are controlled by cis-regulatory elements named imprinting centers (ICs). These elements are 2–4 Kb long DNA sequences characterized by differential DNA methylation levels on the maternal and paternal alleles [3].
The Silver–Russell Syndrome (SRS) is a rare imprinting disorder characterized by slow intrauterine growth and postnatal growth deficiency associated with specific physical characteristics and symptoms [4]. The recent Netchine–Harbison clinical scoring system (NH-CSS) [5] states that clinical diagnosis of SRS can be made in presence of at least four out of the following six criteria: birth weight and/or birth length (≤−2 SD for gestational age), postnatal growth failure (height at 24 ± 1 months ≤−2 SDS or height ≤−2 SDS below mid-parental target height), relative macrocephaly at birth (head circumference at birth ≥1.5 SDS above birth weight and/or length SDS), protruding forehead, body asymmetry (LLD of ≥0.5 cm or arm asymmetry or LLD < 0.5 cm with at least two other asymmetrical body parts) and feeding difficulties (BMI ≤ – 2 SDS at 24 months or current use of a feeding tube or cyproheptadine for appetite stimulation) [4]. Moreover, several other clinical features (e.g. triangular face, clinodactyly, micrognathia, speech delay) are associated with but not specific to SRS [4]. Due to this large spectrum of phenotypes, the clinical diagnosis of this syndrome is challenging. For these reasons, nowadays its incidence is fairly unknown and only estimated to be about 1/30,000–100,000 live births.
The molecular etiology of this disease is unidentified in about 40% of patients. In the remaining 60%, the most frequent abnormalities are epigenetic changes (loss of methylation—LOM) affecting the Imprinting Center 1 (IC1) on chromosome 11p15.5, (30–60% of cases), and maternal uniparental disomy of chromosome 7 (udp(7)mat) (5–10% of patients). Moreover, udp(14)mat and other aberrations at the 14q32.2 imprinted region are reported to be causative of an SRS-like phenotype named Temple syndrome [4, 6].
The imprinted gene cluster on chromosome 11p15.5 is, indeed, organized into two independent domains: the telomeric domain, which includes the growth-repressing noncoding RNA H19 and the growth-promoting IGF2 gene, and the centromeric domain that includes the potassium channel KCNQ1, its antisense KCNQ1OT1, and the growth suppressor CDKN1C gene.
Some CNVs involving the 11p15.5 imprinting cluster have been reported in approximately 1% of SRS cases. The majority of these CNVs correspond to large maternally inherited duplications of 11p15.5 including both the telomeric and centromeric domains, which result in overabundant dosage of the maternally expressed growth suppressor genes [7]. Smaller CNVs have variable phenotypes depending on the number of genes and regulatory elements affected, as well as their parental inheritance, and only a few have been associated with SRS [8,9,10,11,12,13]. Here, we describe a small deletion involving only IC2, which co-segregates with a mild SRS phenotype in a family.
Material and methods
Library preparation and whole-genome sequencing (WGS)
A total amount of 1.0 μg DNA was used as input for sample preparation. Sequencing libraries were generated using NEBNext® DNA Library Prep Kit (New England Biolabs, Ipswich, MA, USA) following manufacturer’s recommendations and indexes were added to each sample. The genomic DNA was randomly fragmented to a size of 350 bp by shearing, then DNA fragments were end-polished, A-tailed, and ligated with the NEBNext adapter for Illumina sequencing (New England Biolabs), and further PCR enriched by P5 and indexed P7 oligos. The PCR products were purified with the AMPure XP system (Beckman Coulter, Milan, Italy) and resulted libraries were analyzed for size distribution by Agilent 2100 Bioanalyzer and quantified using real-time PCR. Libraries were sequenced on an Illumina platform in a 150 bp paired-end mode.
Bioinformatics analysis
Sequencing data were filtered by discarding a read pair if (1) either one read contains adapter contamination; (2) more than 10% of bases are uncertain in either one read; (3) the proportion of low-quality bases is over 50% in either one read. Clean reads were then aligned to the UCSC Genome Browser hg19 reference sequence with the BWA-MEM [14]. BAM file was sorted by SAMtools [15] and duplicates were marked with picard (http://sourceforge.net/projects/picard/). Variant calling and genotyping were performed with the Genome Analysis Toolkit [16]. SNPs and small indels were annotated by ANNOVAR [17] and filtered with NCBI dbSNP v.147, the 1000 Genomes Project catalog (1000g2015aug), and SIFT annotation (dbNSFP version 3.0a) [18]. Structural and copy number variants were identified by Delly [19] and Control-FREEC [20], respectively.
Breakpoint-spanning PCR
To evaluate the correct boundaries of the deletion, a breakpoint-spanning PCR. Primers were designed following data obtained by WGS and purchased from Merk (Merk KGaA, Darmstadt, Germany). PCR products were sequenced by the 3500Dx DNA Sequencer analyzer (Life Technologies) using the BigDye Terminator kit v3.1 (Applied Biosystems). Sequences were aligned with the KCNQ1OT1 reference sequence by MUSCLE v3.8 (https://www.ebi.ac.uk/Tools/msa/muscle/). The deletion has been annotated in the ClinVar database (https://www.ncbi.nlm.nih.gov/clinvar/variation/VCV000977320.1) and in the GV shared LOVD Database (Variant ID: 0000686018, https://databases.lovd.nl/shared/variants/0000686018#00010411).
MS-MLPA
MLPA analysis was performed with the Salsa MS-MLPA Probemix ME030-C3 BWS/RSS (MRC-Holland, Amsterdam, The Netherlands), according to the manufacturer’s protocol in order to detect deletions and duplications and to verify the methylation status of chromosome 11. The kit contains probes evaluating the presence of deletions and duplications in the 11p15.5 genomic region, which includes the following genes: H19, IGF2, KCNQ1, KCNQ1OT1, CDKN1C; and methylation status of imprinting centers, IC1 and IC2 All procedures and data analysis were performed as indicated by the manufacturer. Sizing analysis was performed on a 3500 DX Sequencer (Applied Biosystems, Foster City, CA, USA) using the LIZ 500 Size Standard v2.0. The Coffalyzer.Net software (MRC-Holland) was used to determine fragments length.
Results
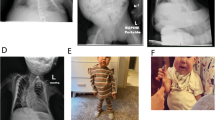
The proband (Fig. 1, III-1) is an 8-year-old girl born from unrelated healthy parents. During prenatal screening, IUGR and oligohydramnios were observed by ultrasound at 27 weeks of gestation. Due to oligohydramnios and flow alteration, urgent cesarean-section delivery was performed at 29 + 5 weeks of gestation. At birth, her weight was 768gr (<3° p, −3SD), height was 31.7 cm (<3° p, −3SD), occipitofrontal circumference was 25.5 cm (10° p, −1SD) and Apgar 5/8. Subsequently, she was hospitalized at the Neonatal Intensive Care Unit for three months, where she was subjected to parenteral feeding and suffered of cholestasis and malabsorption. At 5 months, her parameters were: 3.170 kg weight (<3° p, −3SD), 50.5 cm length (<3° p, −3SD), and 37 cm head circumference (10° p, −1SD). Postnatal follow-up showed slow but regular growth. Physical examination at 8 years showed growth restriction with short stature: weight 15.8 kg (<1° p, −4SD), height 107 cm (<3° p, −3SD). In addition, she had protruding forehead (Fig. 1b), fifth finger clinodactyly, pubarche in absence of axillarche, and apocrine sweating. The Tanner stages were B1 and PH1. Both parents were of normal stature: 170 cm father (II-1) and 160 cm mother (II-2). During genetic counseling, the paternal grandmother (I-1) was referred to be 145 cm tall (<3° p, −3SD), with puberty referred at 8 years old.
At 8 years old, the proband was subjected to surgery for Arnold–Chiari malformation and secondary syringomyelia correction. At 1-year follow-up, she showed a strong syringomyelia reduction. Moreover, growth restriction was confirmed: weight 17.8 kg (<3° p, −3SD), length 110.5 cm (<3° p, −3SDS), BMI 14.5 (10° p, −1SD) and bone age between 7 and 8 years at the Greulich and Pyle scale.
Since the proband was characterized by a generalized growth delay without hemihypotrophy and showed no skeletal or adrenal abnormalities, the NH-CSS was 3/6 (Table 1). Given that clinical SRS diagnosis could be considered when the patient meets at least 4/6 NH-CSS criteria [4], molecular testing for SRS was not performed. Therefore, she was subjected to WGS. The analysis identified a 1.4 Kb deletion in 11p15.5 involving the IC2 region (Fig. 2). To confirm the correct size of the deletion, a breakpoint-spanning PCR was performed. The breakpoints of the deletion were located within the homologous region GGCGCGGG: chr11(GRCh37): g.2,720,674_2,722,054del (Supplementary data 1). Copy number and methylation of IC2 were also investigated by MS-MLPA (Table 2). In the proband, monoallelic deletion was confirmed with 3 out of 4 of the IC2-specific probes; furthermore, approximately 100% level of methylation was detected with all four IC2 probes. These results indicated paternal inheritance of the deletion in III-1, which was indeed confirmed in her father. Moreover, II-1 showed very low IC2 methylation (average 0%), indicating that the deletion was located on his maternal chromosome. This is indeed consistent with his normal growth development. To further characterize this family, we analyzed the proband’s paternal grandmother (I-1) confirming both the presence of the deletion and the high IC2 methylation levels, indicating that the deletion is present on her paternal chromosome. All these data are consistent with the reported growth restriction. Copy number and methylation level of IC1 in all family members were comparable to the controls (Table 2).
Discussion
SRS is a rare condition characterized by growth restriction, peculiar facial features, and body asymmetry. Molecular etiology remains unknown in a substantial fraction of these patients. Indeed, the underlying molecular mechanism is identified only in 40–60% of cases and being mostly represented by loss of IC1 methylation on 11p15.5 or maternal disomy of chromosome 7. Here, we describe a paternal 1.4 Kb deletion of IC2 in a patient with growth delay without hemihypotrophy. The analysis of her clinical data (summarized in Table 1) supported her classification as a mild SRS case.
11p15 microdeletions have been rarely described: maternally inherited deletions affecting the centromeric domain have been described in some cases of Beckwith–Wiedemann Syndrome (BWS) [8,9,10, 13]; while paternally inherited deletions of this region were associated with a few SRS cases. In particular, Gurrieri et al. described a female patient displaying a mild SRS phenotype and harboring a paternal 198 kb deletion on chr11p15.5 that included IC2, KCNQ1OT1 and part of KCNQ1 [21]. More recently, Cytrynbaum et al., described a SRS case harboring a de novo deletion involving part of the KCNQ1OT1 and KCNQ1 genes on the paternally inherited chromosome [11]. The phenotypical comparison of these and our cases are summarized in Table 3.
In our proband, WGS analysis identified a small deletion in 11p15.5 (chr11(GRCh37): g.2,720,674_2,722,054del). The deletion is localized on the paternal chromosome and affects the 5′ end of KCNQ1OT1 and its promoter, IC2 and only a small slice of KCNQ1 intron 11 (Fig. 3). A tiny microdeletion involving 132 bp within the first exon of KCNQ1OT1 and the interval deleted in our case has been recently described and associated with IUGR [12]. Since our proband showed some typical features of SRS in addition to IUGR, it is likely that IC2 deletions of different sizes have different impact on the phenotype.
a Diagram describing the imprinting and methylation levels of the locus. Solid dots in IC1 and IC2 indicates 100% methylation. b Representation of the 1.4 deletion within the 11p15.5 locus, focusing on possible effects on gene expression. Blue striped box: deleted region; light green box: promoter flanking regions; dark green box: minimal promoter; orange box: insulators; golden box: enhancer; black dots: TSS; red dots: MS-MLPA IC2 probes.
Since KCNQ1OT1 is a repressor of the growth suppressor gene CDKN1C, it is expected that deletion of its promoter would activate the expression of this genes and, therefore, cause the occurrence of growth delay. Consistent with the hypothesis of the transcriptional down-regulation of KCNQ1OT1, the most 3′-located MS-MLPA probe shows hypermethylation in the presence of normal copy number. This explanation agrees with data obtained both in animal models and humans. In mice, it was described as the paternal inheritance of a IC2 (also known as Kcnq1ot1 DMR) deletion results in the activation of the maternally expressed genes within the affected domain, which ultimately causes growth deficiency [22]. In humans, a paternal 60 kb deletion encompassing IC2 was described by one of our authors and associated with recurrent severe IUGR [23]. This deletion includes the one observed in the present case, and it results in the loss of KCNQ1OT1 expression and up-regulation of CDKN1C. We previously observed that, due to the presence of multiple distant enhancers, the pathological phenotype is directly influenced by the size of the region involved in the CNV, with larger deletions associated with milder phenotypes [9]. A deletion involving IC2 but sparing all the enhancers on the paternal chromosome, would lead to CDKN1C activation, determining a severe growth restriction. Conversely, deletions also involving one enhancer or more would result in a weaker or no activation of paternal CDKN1C, causing milder phenotypes [24].
It should be acknowledged that the 1.4 kb deletion assessed in the present case only partially involves the 600 bp KCNQ1OT1 minimal promoter (MP) (Fig. 3). Indeed, the FANTOM 5.0 database (https://fantom.gsc.riken.jp/zenbu) identified at least seven alternative transcriptional start site (TSS) for KCNQ1OT1 within the MP. Furthermore, this chromosome region is filled with regulatory elements (i.e. enhancers, insulators, and silencers), whose interactions and tissue specificity are still to be elucidated [22, 25]. Our deletion impairs the core promoter region sparing both alternative TSS and enhancer sequences (Fig. 3b). For this reason, it is possible that the phenotype of the present case that is less severe than the one associated with the 60 kb deletion is due to residual regulatory features controlling KCNQ1OT1, impairing but not completely abolishing the repressing activity on the CDNK1C growth suppressor gene. KCNQ1OT1 is not expressed on the maternal chromosome due to methylation of its promoter. This likely explains why paternal but not maternal inheritance of the 1.4 kb deletion alters the expression of growth restraining genes and results in a growth deficiency phenotype in our family. Accordingly, maternal deletion of the homologous mouse sequence has no effect on imprinted gene expression or growth [22].
References
Ishida M, Moore GE. The role of imprinted genes in humans. Mol Asp Med. 2013;34:826–40.
Monk D, Mackay DJG, Eggermann T, Maher ER, Riccio A. Genomic imprinting disorders: lessons on how genome, epigenome and environment interact. Nat Rev Genet. 2019;20:235–48.
Boonen SE, Freschi A, Christensen R, Valente FM, Lildballe DL, Perone L, et al. Two maternal duplications involving the CDKN1C gene are associated with contrasting growth phenotypes. Clin Epigenetics. 2016;8:69.
Wakeling EL, Brioude F, Lokulo-Sodipe O, O’Connell SM, Salem J, Bliek J, et al. Diagnosis and management of Silver–Russell syndrome: first international consensus statement. Nat Rev Endocrinol. 2017;13:105–24.
Azzi S, Salem J, Thibaud N, Chantot-Bastaraud S, Lieber E, Netchine I, et al. A prospective study validating a clinical scoring system and demonstrating phenotypical-genotypical correlations in Silver-Russell syndrome. J Med Genet. 2015;52:446–53.
Kagami M, Nagasaki K, Kosaki R, Horikawa R, Naiki Y, Saitoh S, et al. Temple syndrome: comprehensive molecular and clinical findings in 32 Japanese patients. Genet Med. 2017;19:1356–66.
Brown LA, Rupps R, Peñaherrera MS, Robinson WP, Patel MS, Eydoux P, et al. A cryptic familial rearrangement of 11p15.5, involving both imprinting centers, in a family with a history of short stature. Am J Med Genet A. 2014;164:1587–94.
Niemitz EL, DeBaun MR, Fallon J, Murakami K, Kugoh H, Oshimura M, et al. Microdeletion of LIT1 in Familial Beckwith-Wiedemann Syndrome. Am J Hum Genet. 2004;75:844–9.
Algar E, Dagar V, Sebaj M, Pachter N. An 11p15 imprinting centre region 2 deletion in a family with beckwith wiedemann syndrome provides insights into imprinting control at CDKN1C. PLoS ONE. 2011;19:6:e29034.
Baskin B, Choufani S, Chen Y, Shuman C, Parkinson N, Lemyre E, et al. High frequency of copy number variations (CNVs) in the chromosome 11p15 region in patients with Beckwith–Wiedemann syndrome. Hum Genet. 2014;133:321–30.
Cytrynbaum C, Chong K, Hannig V, Choufani S, Shuman C, Steele L, et al. Genomic imbalance in the centromeric 11p15 imprinting center in three families: Further evidence of a role for IC2 as a cause of Russell–Silver syndrome. Am J Med Genet A. 2016;170:2731–9.
Eggermann T, Kraft F, Lausberg E, Ergezinger K, Kunstmann E. Paternal 132 bp deletion affecting KCNQ1OT1 in 11p15.5 is associated with growth retardation but does not affect imprinting. J Med Genet. https://jmg.bmj.com/content/early/2020/05/23/jmedgenet-2020-106868. 2020.
Zollino M, Orteschi D, Marangi G, Crescenzo AD, Pecile V, Riccio A, et al. A case of Beckwith–Wiedemann syndrome caused by a cryptic 11p15 deletion encompassing the centromeric imprinted domain of the BWS locus. J Med Genet. 2010;47:429–32.
Li H, Durbin R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinforma Oxf Engl. 2009;25:1754–60.
Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, et al. The sequence alignment/map format and SAMtools. Bioinforma Oxf Engl. 2009;25:2078–9.
DePristo MA, Banks E, Poplin R, Garimella KV, Maguire JR, Hartl C, et al. A framework for variation discovery and genotyping using next-generation DNA sequencing data. Nat Genet. 2011;43:491–8.
Wang K, Li M, Hakonarson H. ANNOVAR: functional annotation of genetic variants from high-throughput sequencing data. Nucleic Acids Res. 2010;38:e164.
Liu X, Jian X, Boerwinkle E. dbNSFP: a lightweight database of human nonsynonymous SNPs and their functional predictions. Hum Mutat. 2011;32:894–9.
Rausch T, Zichner T, Schlattl A, Stütz AM, Benes V, Korbel JO. DELLY: structural variant discovery by integrated paired-end and split-read analysis. Bioinforma Oxf Engl. 2012;28:i333–9.
Boeva V, Popova T, Bleakley K, Chiche P, Cappo J, Schleiermacher G, et al. Control-FREEC: a tool for assessing copy number and allelic content using next-generation sequencing data. Bioinforma Oxf Engl. 2012;28:423–5.
Gurrieri F, Zollino M, Oliva A, Pascali V, Orteschi D, Pietrobono R, et al. Mild Beckwith-Wiedemann and severe long-QT syndrome due to deletion of the imprinting center 2 on chromosome 11p. Eur J Hum Genet. 2013;21:965–9.
Fitzpatrick GV, Soloway PD, Higgins MJ. Regional loss of imprinting and growth deficiency in mice with a targeted deletion of KvDMR1. Nat Genet. 2002;32:426–31.
De Crescenzo A, Sparago A, Cerrato F, Palumbo O, Carella M, Miceli M, et al. Paternal deletion of the 11p15.5 centromeric-imprinting control region is associated with alteration of imprinted gene expression and recurrent severe intrauterine growth restriction. J Med Genet. 2013;50:99–103.
Cerrato F, De Crescenzo A, Riccio A. Looking for CDKN1C enhancers. Eur J Hum Genet. 2014;22:442–3.
Schultz BM, Gallicio GA, Cesaroni M, Lupey LN, Engel N. Enhancers compete with a long non-coding RNA for regulation of the Kcnq1 domain. Nucleic Acids Res. 2015;43:745–59.
Acknowledgements
The authors thank the patient and her family for participating in this study.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
About this article
Cite this article
Mio, C., Allegri, L., Passon, N. et al. A paternally inherited 1.4 kb deletion of the 11p15.5 imprinting center 2 is associated with a mild familial Silver–Russell syndrome phenotype. Eur J Hum Genet 29, 447–454 (2021). https://doi.org/10.1038/s41431-020-00753-1
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41431-020-00753-1