Abstract
Sludge biosolids for agricultural application represent a valuable fertilizer but also a health risk unless pathogens are effectively reduced, and recontamination controlled. The Post Anaerobic Digestion Thermal Hydrolysis Process (Post-AD THP) is gaining interest due to improved dewaterability, reducing the volume and thus transportation costs of biosolids. However, Post-AD THP results in sterile biosolids easily exposed to recontamination by pathogens due to the lack of microbial competitors. In theory, this could be suppressed by establishing a competing community of harmless bacteria. The theory was tested by monitoring the abundance of Escherichia coli (viable counts) and gene abundance (ddPCR) in wastewater recontaminated Post-AD THP biosolids, with and without addition of compost. Respiration, total bacterial population and bacterial diversity (16S rRNA amplicon sequencing) were used to monitor the microbial community. Biosolids from the regulatory approved methods thermophilic AD (TAD) and Pre-AD THP were tested in parallel for comparison. The results demonstrated that regulatory requirements can be reached by storing the TAD and Pre-AD THP biosolids for 3 days after recontamination and the Post-AD THP biosolids for more than 13 days. However, addition of compost suppressed growth of E. coli in Post-AD THP biosolids, reducing the time to comply with regulative requirements. In conclusion, pathogen growth in Post-AD THP biosolids can be controlled by inoculation with compost.
Graphical abstract
Similar content being viewed by others
Statement of Novelty
Safe management and pathogen control of sludge biosolids is essential for its further value as a fertilizer on agricultural land. Indicator bacteria growth after conventional anaerobic digestion (AD) configurations have been thoroughly studied in current literature. However, introduction of new technology such as the post AD thermal hydrolysis process (Post-AD THP) challenges the existing body of knowledge and legislations in some countries. Legislations must distinguish between inadequate sanitation in the sludge treatment facility and growth of pathogens due to recontamination during storage and handling. To the best of our knowledge, growth of pathogens due to recontamination in Post-AD THP biosolids has so far not been studied and will provide important input for further research and development of government regulations.
Introduction
Treatment and stabilization of sewage sludge is needed before safe land application so that it can be valorized as a fertilizer rich in nutrients and organic matter without transmitting infectious disease. This can be done through anaerobic digestion (AD) at mesophilic or thermophilic temperatures producing a solid residue often termed biosolids [1]. While biosolids are produced continuously throughout the year, the period for land application is limited and season dependent. Consequently, the biosolids must be stored depending on the season and regulatory requirements from a few days to several months until use. Ensuring safe handling therefore requires knowledge on the risk for regrowth of pathogens in biosolids generated with different sanitation methods, but also the biosolids’ resistance to external recontamination during storage. Previous studies have documented indicator bacteria survival and regrowth of conventional systems [2] with E. coli increasing up to 108–109 cells/g dry solids (DS) during the first storage days [3]. However, with new sludge treatment processes and sanitation methods, pathogen growth due to recontamination challenges the existing literature and legislation framework and must be investigated in greater depth.
Local legislation usually regulates safe biosolids’ use as fertilizers by setting limits for fecal coliforms, Salmonella, Helminth ova and enteric viruses [4]. However, the limits differ between countries. For instance, the limit for fecal coliforms (Most Probable Number; MPN) in Norway and the USA is 2500 and 1000 MPN/DS, respectively, and for Salmonella sp. 0 and 0.75 MPN/DS, respectively [1, 5]. To follow the legislations of safe biosolids, the most commonly used indicator group is thermotolerant coliform bacteria (TCB) and the more specific bacterium Escherichia coli (E. coli) [3, 6, 7] since it is present in much larger numbers than Salmonella [8]. The amount of indicator organisms has traditionally been assessed by viable counts (MPN), but this has been contested [8] because fecal coliforms and E. coli may enter a “viable but non-culturable state” during digestion [3]. Therefore, researchers have used both MPN and polymerase chain reaction (PCR) to identify fecal regrowth in centrifuged biosolids [3, 7, 9]. In addition to pathogen reduction, it is important to reduce the attractiveness of the biosolids for vectors such as flies, insects or birds. These vectors, can serve as recontamination routes of the treated biosolids [10]. Specific legislations such as the Vector Attraction Reduction (VAR) requirements in the US [1] have been introduced as a consequence of reports on recontamination and growth of pathogens in sterilized biomass [11].
Several methods are in use to meet the regulations for both vector attraction and pathogen reduction. For example, the presence of diverse groups of microorganisms has shown to compete and successfully suppress the growth of pathogens such as Salmonella [10, 12]. However, acceptable levels of indicator bacteria are normally obtained by thermal drying, thermophilic anaerobic digestion (TAD) in batch or multi-stage, mesophilic AD in series, pasteurization, conditioning with lime or the thermal hydrolysis process (THP) [4, 13, 14]. In addition, the biosolids storage environment may affect pathogen survival [15].
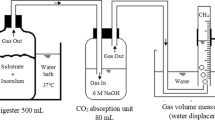
THP effectively sterilizes the material (165 °C) and has traditionally been applied before the AD process (Pre-AD THP, Fig. 1) [16] meeting the highest standard for biosolids hygienization and the VAR requirements. However, recently the THP process has been applied after AD (Post-AD THP, Fig. 1), where digestate is dewatered immediately after THP treatment and the centrate containing soluble and easily degradable organic material is recirculated to the digester (Fig. 1) improving both biogas production and dewaterability, significantly reducing the wet biosolids mass for disposal [17, 18]. While this treatment process ensures absence of all pathogens and no regrowth, it also eliminates all other microbes, introducing a potential for recolonization by pathogens. Hence, these biosolids are uncompliant with the VAR requirements in the US unless it is dried to >75% DS. In the current Norwegian legislation, such biosolids are not specified. The degree of recolonization is however unknown, as to our best knowledge there is no available literature on pathogen growth in Post-AD THP biosolids. In addition, an alternative to drying that needs to be explored is inoculating the biosolids with harmless microbes immediately after the Post-AD THP treatment and dewatering as a part of the handling and storage facility, replacing the typical inoculation taking place in AD.
The overall objective of this study was therefore to investigate if inoculating the dewatered Post-AD THP biosolids with a mixture of harmless bacteria hinder growth of E. coli after recontamination. For comparison, the study included two biosolids approved by Norwegian legislation (TAD and Pre-AD THP) [5] giving a total of four cases to compare.
To achieve the objective, the effect of inoculating Post-AD THP treated biosolids with pathogen-free compost on the subsequent E. coli growth was tested. The source of E. coli was wastewater, added 2 days after the compost inoculation or 2 days storage for Post-AD THP biosolids without compost, Pre-AD THP biosolids and TAD biosolids. The MPN method and digital droplet PCR (ddPCR, primers targeting the uidA gene in E. coli) were used to monitor the growth of E. coli. In addition to the commonly used MPN and ddPCR methods three more methods were used to investigate the hypothesis: (1) a robotized incubation system to monitor the microbial respiration, (2) ddPCR of 16S rRNA to assess the total bacterial population, and (3) bioinformatics analysis of 16S rRNA to investigate the microbial diversity of the biosolids before and after recontamination, in addition to the effect of compost addition to the Post-AD THP biosolids.
Materials and Methods
Samples
Plant Sampling and Compost
Three biogas plants with different hygienization methods were selected for analysis (Table 1). Dewatered biosolids were collected at the outlet of the dewatering equipment and placed in airtight containers at Plant A and B at the same day and stored overnight at 4 °C before the experiments were started. Dewatered Post-AD THP biosolids were collected at the centrifuge outlet at Plant C in airtight containers, shipped to the Norwegian University of Life Sciences and stored at 4 °C until the experiment was started. To ensure sterile biosolids after sampling and transportation from Plant C, these biosolids were autoclaved at 121 °C for 15 min prior to the experiment. The autoclave treatment was assumed not to alter the sample characteristics significantly since the sludge had undergone prior THP treatment at higher temperature.
Dewatering liquor was collected at all three plants to measure the soluble chemical oxygen demand of the liquid phase of the biosolids.
The sterile biosolids sample (Post-AD THP) was split in two. One part was blended with 10 vol% compost (w/w) by hand and the other was used directly. Compost was provided by the Norwegian company Høst. According to manufacturer the compost was based on yard waste, processed at 60–70 °C in 2–3 months with frequently turning and air supply, and subsequent vermicomposting. The compost was documented free of fecal coliforms and E. coli by enumeration according to NS-EN ISO 9308-2:2014. The compost was stored at 4 °C and left at room temperature the last 24 h before start-up.
All samples were passed through a sieve with a mesh size of 4 mm to remove clumps and large particles, to secure uniform and representative subsamples. The sieve was washed and sanitized by a 70 vol% ethanol solution between samples. In total, four sample sets were prepared for analysis. All samples were gently homogenized by hand prior to sample preparation.
Source of Recontamination
Wastewater collected from the primary sedimentation tank at Plant B was used as the source of E. coli. The sample was transferred to the laboratory in an airtight container and stored at 20 °C until use. Presence of E. coli in the sample was confirmed by enumeration by a commercial laboratory prior to recontamination of samples sets.
Robotized Incubation System for Respiration Kinetics
To investigate the respiration rates of the microbial community with and without recontamination with wastewater, a robotized incubation system connected to a gas chromatograph (7890A, Agilent Technologies) and a NO analyzer (Model 200E Chemilumenescence NO analyzer, Teledyne) previously described by Molstad et al. [19] was used. In brief: the system allows for automated time incremental sampling of the headspace of up to 42 serum vials, held in a thermostated water bath, thus monitoring the kinetics of O2, N2, NO, N2O, CO2 and CH4. The sample volume used for analysis is replaced by helium. Six sterilized serum vials (120 mL) were used per sample set (recontaminated + control). 2 g of biosolids were added to each vial, capped with an airtight rubber septum, and incubated aerobically at 20 °C. Mass loss of gasses due to sampling and dilution with helium was corrected for.
After 48 h three flasks were recontaminated by spiking with 50 µL wastewater, while the other three replicate flasks received equal amounts of sterile distilled water by a syringe. To secure aerobic conditions, O2 was injected when needed to keep the O2 concentration in the headspace within the interval 10–20 vol%. The molar ratio of O2 consumption to CO2 production was close to 1 (Supplementary Material, Fig. S6), indicative of aerobic respiration throughout [20]. CO2 was consequently chosen to display the respiratory data as it was less affected by injection of O2 (the rate of O2 consumption could not be calculated for those time intervals when O2 was injected).
Preparation of Samples
In parallel with the incubation robot experiment, a large set of samples with identical treatments, destined for destructive sampling throughout, to determine the viable counts and gene abundances was incubated. For the samples to be recontaminated with wastewater, triplicates for each sampling day were prepared in 50 mL sample tubes by adding 23 g sample to each tube at the start of the experiment. This resulted in 39 tubes per treatment. For the control samples, that were not recontaminated with wastewater, duplicates of 700 mg sample for each sampling day were prepared in 2 mL cryogenic tubes at the start of the experiment. The samples were all placed in closed plastic boxes at room temperature (20 °C) with the cap loose to allow air diffusion into the tube. Each sample set was placed in separate containers and the bottom was covered with tissues wetted with distilled water to avoid desiccation of the samples. The plastic containers were opened for recontamination and sampling, otherwise left closed. At each time of sampling, two tubes were frozen (−80 °C) for ddPCR and 16S rRNA gene sequencing, and one was transported to a commercial laboratory in a cooler (~4 °C) for enumeration of E. coli.
The compost and the sterile biosolids (Post-AD THP) were mixed gently by hand 48 h prior to recontaminating with wastewater, to give the compost bacteria a reasonable head-start. 48 h typically represent the storage capacity of receiving containers at wastewater treatment plants to reduce work hours in weekends, hence simulating the maturing time available in full-scale systems. After 48 h recontamination was induced by distributing 0.5 mL of wastewater across the top layer of the samples. The volume of wastewater contamination was based on initial investigations with the incubation robot (data not shown). The tubes were capped, shaken and left with the cap loosely placed on top of the tube.
The dry solids concentration of each sample-set was monitored from day −3 to day 9 by drying a sample at 105 °C overnight (Supplementary Material, Fig. S8).
Microbial Analysis
Enumeration of E. coli by Standard Cultivating Methods
Enumeration of E. coli and total fecal coliforms were done by an accredited commercial laboratory according to NS-EN ISO 9308-2:2014 to assess the viable count of E. coli and total fecal coliforms.
DNA Extraction and Purification
Genomic DNA from duplicate samples from each sample time point was extracted using the DNeasy PowerSoil Kit (Qiagen, Germany). The entire process was carried out according to the manufacturer’s protocol. The bead beating was performed with a FastPrep 24 at 4 m/s and 45 s.
Enumeration of E. coli by Digital PCR
ddPCR was performed on both technical replicate DNA extracts. The ddPCR reaction mix contained 10 µL QX200 ddPCR EvaGreen Supermix (Bio-Rad), 2 µL of DNA template/extract, and 100 nM final concentration of primer pairs PRK341F/PRK806R (5′-CCTACGGGRBGCASCAG-3′, 5′-GGACTACYVGGGTATCT-3′) (Eurofins Genomic) targeting the V3-V4 hypervariable region of prokaryotic 16S rDNA [21], or primer pairs ECF_uidA/ECR_uidA (5′-CGGAAGCAACGCGTAAACTC-3′, 5′-TGAGCGTCGCAGAACATTACA-3′) (Eurofins) targeting a region of the beta-glucuronidase reporter gene (uidA) specific for E. coli [22] (Table 2). 20 µL reaction mix and 70 µL Droplet generation oil for EvaGreen (Bio-Rad) was used to generate droplets in a QX200 droplet generator (Bio-rad). 40 µL of oil droplet suspension was transferred to 96 well twin.tec PCR plates (Eppendorf) and heat sealed with aluminium foil [PX1™ PCR plate sealer (Bio-Rad)].
PCR cycling conditions (Table 3) when amplifying uidA primer products (90 bp) were in accordance with the QX200™ ddPCR™ EvaGreen Supermix PCR protocol with an annealing/extension temperature of 63 °C. The cycling conditions when amplifying 16S rDNA (465 bp) were 95 °C for 5 min (enzyme activation/denaturation), 40 cycles at 95 °C for 30 s (denaturation) and 55 °C for 30 s (annealing/extension) and 45 s at 72 °C (extension), followed by signal stabilization for 5 min at 4 °C and 5 min at 90 °C. All PCR reactions were conducted in a 2720 Thermal Cycler (Applied Biosystems) with 2 °C s−1 ramp rate, a lid temperature of 105 °C and a final 4 °C hold step. PCR products were analyzed in a QX200 droplet reader (Bio-Rad), and the data was analyzed in the Quantasoft™ Analysis Pro 1.0.596 software (Bio-Rad).
16S rRNA Gene Sequencing and Bioinformatics Analysis
To assess taxonomic composition and dynamics of the prokaryotic community, the 16S rDNA amplicons of the extracted DNA were sequenced. For this, a two-step nested PCR was carried out, first amplifying the V3–V4 region using the primer pair PRK341F(5′-CCTACGGGRBGCASCAG-3′)/PRK806R(5′-GGACTACYVGGGTATCTAAT-3′) [21], followed by a second PCR for attachment of dual indices and Illumina sequencing adaptors. Both PCRs were performed using 1 × HotFirePol blend master mix (Solis BioDyne), and amplicons were purified with AMPure XP beads (Beckman-Coulter, USA) using the manufactures protocol. Equimolar concentrations of the amplicon libraries were pooled and the final library was quantified with the QX200™ Droplet Digital™ PCR System (Bio-Rad, USA) using primers targeting the Illumina adapters. Finally, the library was sequenced on an Illumina MiSeq system (Illumina, USA), using the MiSeq v3 reagent kit with 300-bp paired-end reads.
The resulting sequences were processed using USEARCH associated algorithms. Initially, paired end reads were merged, filtered (maxee = 1, minimum length = 350 bp), dereplicated and singletons discarded. Sequences were then clustered to operational taxonomic units OTUs at 97% identity threshold and checked for chimeric sequences. Finally, taxonomy was assigned through the uclust method implemented in QIIME [23], using the SILVIA database [24]. UniFrac distances (weighted and unweighted) were generated through the beta_diversity.py script, also in QIIME. Samples with low sequencing depth, i.e. below 10,000 sequencing reads per sample (in total 7 samples, see Supplementary Material, Fig. S12), were excluded from the downstream analysis. The generated OTU data was mined and statistically analyzed and visualized using Calypso [25]. Taxa less than 0.01% relative abundance across all samples have been filtered out from the final diversity plots, and all analysis are based on total sum normalization (TSS) + square root transformed data, unless stated otherwise.
Results and Discussion
Growth of E. coli
Growth of E. coli in the four sample-sets was monitored by gene abundance (ddPCR of the uidA gene) (Fig. 2a) and by viable counts (Fig. 2b). The strictest regulatory requirement for biosolids in Norway is less than 2500 viable fecal coliforms (MPN/gDS) [5] and in the US the limit is 1000 total fecal coliforms (MPN/gDS) [1]. Both E. coli and total fecal coliforms were enumerated, and the two were practically identical throughout the entire incubation (Supplementary Material, Fig. S1). Data for each sample-set separately can be found in Supplementary Material Fig. S3–S5.
Density of E. coli throughout the incubation: abundance of uidA gene quantified by ddPCR (a) and viable counts (MPN) (b), both plotted against time. TAD thermophilic anaerobic digestion, Post-AD THP biosolids sterilized by THP after AD, Pre-AD THP biosolids sterilized by THP prior to AD. All biosolids were contaminated by wastewater immediately after sampling at time 0 (marked by arrow). USEPA limit of 1000 MPN/gDS is indicated by the black line. It should be noted that the very high MPN values for Post-AD THP at day 1 and 3 are minimum estimates (all MPN tubes were positive). The detection limit for the MPN method is indicated by striped area (all MPN tubes negative), which implies that all viable counts prior to wastewater contamination were below the detection limit. The same is true for all viable counts after day 7, except for Post-AD THP
The Post-AD THP treatment resulted in viable counts below detection limit (Fig. 2b) but had not destroyed the E. coli uidA gene (Fig. 2a) and hence the abundance of E. coli uidA gene (Fig. 2a) was 14,908 copies/gDS in the Post-AD THP biosolids without compost. It showed no immediate increase by the recontamination, but increased gradually (1–2 orders of magnitude) during the first 2 days after recontamination, remaining high throughout the rest of the incubation. The viable counts of E. coli (Fig. 2b) were below the detection limit before recontamination, spiked to high numbers in response to contamination, fluctuated throughout the next days and declining to low levels after 13 days (Fig. 2b). Adding compost to this biosolids (Post-AD THP + Com) evidently suppressed E. coli: the uidA gene abundance remained low compared to that without compost (Fig. 2a) and the viable counts declined rapidly to very low values after the first spike (Fig. 2b).
The addition of compost to the Post-AD THP biosolids was effective in reducing the time for E. coli viable counts to reach below limit for the Norwegian and US regulatory requirements (Fig. 2b). In the sterile Post-AD THP biosolids, it took 13 days for the viable counts of E. coli to reach below the US limit, but only 4–8 days if compost had been added 2 days before recontamination. Norwegian regulations were met after 13 days of storage for Post-AD THP biosolids, and 3–4 days with the addition of compost. Regulatory requirements can therefore be fulfilled either by allowing storage times above 13 days or by adding compost to reduce the time and space needed for storage. This is in line with work by other researchers showing that substrate- and oxygen availability stimulate some growth of fecal coliforms, but that low levels are reached within two weeks [7].
The Post-AD-THP biosolids with compost performed almost as well as the Norwegian currently accepted method TAD (Thermophilic digestion without THP). In the biosolids that were sterilized before anaerobic digestion (Pre-AD THP), viable count of E. coli peaked at day 2 just above the USEPA limit and 3 days of storage are needed to comply with US regulations, however, the peak was lower than that for TAD biosolids. Pre-AD THP never exceeded the Norwegian regulation limit after recontamination, while the TAD biosolids did on day 1 and 2.
In TAD biosolids the viable counts of E. coli were low before contamination and from day 3, while uidA abundance remained much higher than in any of the others. This could be due to differences in the amount of extracted DNA between samples but more likely related to the lower treatment temperature these biosolids have experienced (55 °C) compared to the others (165 °C). The latter can be supported by the findings of Kirkegaard et al. [26] where no overlap between the bacterial community of the digester feed sludge and digestates in plants with THP was detected, in contrast to both mesophilic and thermophilic digestates without THP.
As substrate availability has been linked to pathogen growth in biosolids [7], the soluble organic material in the biosolids was measured as chemical oxygen demand (COD) in the dewatering liquor of TAD, Pre-AD THP and Post-AD THP. The soluble COD was used as surrogate measurement to assess the available substrate for microbial growth. As expected, the Post-AD THP had the highest soluble COD concentration (32.3 ± 0.6 mg COD/L, data not shown) due to the thermal hydrolysis treatment immediately before dewatering. TAD had the second largest soluble COD concentration (10.7 ± 0.2 mg COD/L, data not shown) followed by the Pre-AD THP (3.5 ± 0.1 mg COD/L, data not shown). The extent of E. coli growth in the biosolids tested in this study can be linked to the concentration of soluble COD in the dewatering liquor. Hence, these results are in line with findings from Higgins et al. [3] and Chen et al. [7]. The faster decline of culturable E. coli in Post-AD THP + Com compared to Post-AD THP could therefore be due to the competition for substrate with the compost microbial community and subsequent substrate depletion. The fluctuating levels of culturable E. coli for Post-AD THP during the 13-day period (Fig. 2b) could be due to the alternation between different carbon feed sources, and the adaptation of the bacterial community to these substrates.
Viable counts and gene abundance of E. coli showed that compost inoculation was efficient in suppressing E. coli growth in sterile Post-AD THP biosolids, and the 13 days of storage could be reduced to 8. Increased microbial activity due to recontamination can also be measured as increased respiration rates. To investigate this a robotized incubation system was used.
Respiration
The rates of CO2 production throughout the incubation period are shown in Fig. 3. The O2 consumption rates were essentially replicas of these figures because the molar respiratory quotient (RQ = O2-consumption/CO2-production) was ~1 throughout the entire incubation of all biosolids, except for an initially RQ in Pre-AD THP (first 10–20 h), as shown in Supplementary Material, Fig. S6.
Respiration kinetics. The panels show the average respiration rates (µmol CO2 g−1 dry solids h−1) in the different biosolids, recontaminated with wastewater (at time 0), and their respective controls (without wastewater contamination): Post-AD THP (a), Post-AD THP + Compost (b), Pre-AD THP (c) and TAD (d)
The Post-AD THP biosolids in Fig. 3a with no wastewater should in principle be sterile, hence without any respiration. Nevertheless, low but significant rates of O2 consumption (Supplementary Material, Fig. S6) could be detected. This could be due to abiotic reactions, or a combination of abiotic reactions and low biological activity by organisms introduced with the distilled water added at time 0. After the sterile Post-AD THP biosolids were contaminated with wastewater (Fig. 3a), a conspicuous series of respiratory spikes were detected at day 1, 3, 5 and 7 [O2 consumption corresponded with the spikes in exactly the same way (Supplementary Material, Fig. S6B)], suggesting microbial respiration. These conspicuous spikes may be explained by a shift in cellular metabolism upon depletion of specific carbon sources of dominating organisms that in turn determine the lag duration [27], or, that cells introduced by the wastewater exploited different fractions of available carbon and reached high cell densities at different timepoints depending on the initial number of organisms in each fraction.
Adding compost to the Post-AD THP biosolids (Post-AD THP + Com, Fig. 3b) had a striking effect on the respiration: it was initially low, but increased gradually to peak after 3 days, but remained high throughout compared to that with Post-AD THP biosolids (Fig. 3b vs. a). Adding wastewater in addition to compost had negligible effects on the respiration, except for a stronger spiking of the respiration after 3 days.
The biosolids that had been THP treated prior to AD (Pre-AD THP, Fig. 3c) had low respiration rates throughout, peaking at day 2. Adding wastewater to these biosolids enhanced the respiration spike but had otherwise no significant effect on the respiration. A slight increase in respiration from day 9 can be observed and could be related to the growth of prokaryotes (Fig. 4).
The biosolids that had not been THP treated at all (TAD, Fig. 3d) had a similar respiration kinetics as Pre-AD THP (Fig. 3c), but higher rates throughout, and a stronger peak after 2 days. The addition of wastewater had marginal effects apart from a slightly stronger respiration peak after 2 days.
Overall, the response in respiration when adding wastewater in Post-AD THP + Com (Fig. 3b), Pre-AD THP (Fig. 3c) and TAD (Fig. 3d) biosolids are similar, while Post-AD THP biosolids differ from the others.
The incubation was continued for 25 days (Supplementary Material, Fig. S6 and S7), and the cumulated CO2 at this time point was used to compare the fraction of total organic carbon mineralized in the various biosolids, depending on the organisms’ present. The results (Supplementary Material, Table S1) show that after 25 days, the organisms in the compost mineralized 8–9% of the total C in Post-AD THP, while the organisms in the wastewater mineralized only 2–3%. The indigenous organisms in the Pre-AD THP mineralized 6–7% of total C (recontamination with wastewater had a marginal effect on this), while the indigenous organisms in TAD mineralized 9–10%.
Taken together, these results demonstrate that the three biosolids contain a significant fraction of degradable organic material that can sustain respiratory metabolism and growth, and that organisms in the compost evidently have a capacity to utilize more of these compounds than the organisms in wastewater (including E. coli). It appears likely, therefore, that inoculation with compost would minimize the potential of growth of pathogens by scavenging the available carbon substrates. Any antagonistic effects would add to the suppression of pathogens.
The respiration trials support the observations of E. coli growth in Fig. 2b, showing highest activity in the first days for Post-AD THP + Com, Pre-AD THP and TAD and fluctuating and prolonged activity in the Post-AD THP biosolids. Microbial respiration in the sterile Post-AD THP biosolids had a final peak around day 13, afterwards respiration remained constant and low throughout the test period (25 days, Supplementary Material, Fig. S6B), supporting the previous results that 13 days of storage could be sufficient. Post-AD THP + Com showed stable and low respiration rates after the main peak ending after 3–4 days (Fig. 3b) and until day 25 (Supplementary Material, Fig. S7) supporting the hypothesis that inoculation with compost will stabilize the Post-AD THP biosolids faster. However, the observations could also be due to growth of other microorganisms in addition to E. coli and the growth of total prokaryotes where therefore investigated.
Growth of Prokaryotes (16S rRNA Genes)
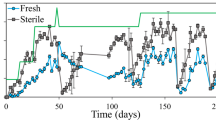
Total 16S rRNA genes in DNA extracts were quantified (Fig. 4) with ddPCR to assess growth of prokaryotes and to adjust for bias in DNA extraction when interpreting uidA gene copies. As expected, the 16S rRNA gene abundance was low initially in sterile Post-AD THP biosolids but increasing (albeit fluctuating) after recontamination (at day 0), presumably by growth. The 16S rRNA gene abundance was significantly higher in the TAD biosolids compared to the others.
No correlation between increased respiration and increased 16S rRNA gene abundance was found. In theory, the spikes of respiration (Fig. 3) should reflect high growth rates, hence increase in the 16S rRNA gene abundance. However, it seems likely that their net growth is balanced by death of others, possibly by protozoan grazing [28, 29]. The only apparent exception is the early net increase in 16S rRNA genes after recontamination of Post-AD THP biosolids without compost, which coincided with the first respiration spike (Fig. 3a). Inspection of the growth yield per mol O2 for this period showed a growth yield of 1.2 × 1014 16S rRNA gene copies mol−1 O2, in comparison the determined oxic growth yield of the model bacterium Paracoccus denitrificans is 1.5 × 1014 cells mol−1 [30]. Hence, we consider the relative increase in the 16S rRNA gene to be reliable for determining growth of the total prokaryotic population.
Biases due to inadequate DNA extraction with commercial kits make it challenging to compare different types of samples. The ratio of uidA copies to the prokaryotic population (16S rRNA) was therefore calculated to obtain less biased estimates of the relative abundance of E. coli in the different biosolids throughout the incubation.
Escherichia coli (uidA) Compared to Prokaryotic Population (16S rRNA)
The ratio of uidA copies to 16S rRNA was plotted against time in Fig. 5 to assess the relative abundance of E. coli in the total population during the incubation period with contaminated samples. The uidA/16S rRNA of control samples (without contamination) can be found in Supplementary Material, Fig. S11.
The uidA/16S rRNA abundance ratio was <12 × 10–6 throughout the entire incubation, hence, E. coli constituted a marginal fraction of the total microbial population in all biosolids.
In the Post-AD THP biosolids without compost, the uidA/16S rRNA ratio increased rapidly during the first two days after contamination with wastewater (Fig. 5), coinciding with the first respiration peak in these biosolids (Fig. 3a). This suggests that when E. coli has no other competitors than those present in the wastewater, it grows faster than the average of the entire population. However, the uidA/16S rRNA declined gradually throughout the subsequent 5 days (days 2–7), coinciding with the next two spikes of respiration (Fig. 3a). As noted previously, the spiking of respiration in these biosolids was interpreted as a succession of different groups of organisms. It is tempting to speculate that the second and third peak of respiration is driven by organisms that suppress E. coli, either by antagonistic effects or by competing for carbon substrates. Nevertheless, the uidA/16S rRNA ratio declined approaching similar levels as before recontamination well within 13 days supporting observations in Fig. 2 that this would be a sufficient timeframe for reduction of E. coli after recontamination.
The effect of adding compost to the Post-AD THP biosolids was striking but must be interpreted with care: prior to the contamination with wastewater (time < 0), the uidA/16S rRNA ratio was ~10 times lower than that without compost. This is attributable to dilution with compost bacteria (total 16S rRNA was approximately 10 times higher with than without compost, Fig. 4). Recontamination with wastewater increased the uidA/16S rRNA, but it remained low except for a spike at day 4. This spike could suggest a late proliferation of E. coli, but the absolute abundance of uidA (Fig. 2a) lends no support to this. A more likely explanation is that a fraction of the bacteria introduced with the compost died out at this point in time, seen as a decline in 16S rRNA abundance (Fig. 4).
The results confirm that addition of compost is an efficient strategy in reducing the growth of E. coli compared to the total microbial population in Post-AD THP biosolids. Pre-AD THP biosolids remained unchanged by recontamination. TAD had a higher abundance of E. coli genes, also when normalized to the total microbial population throughout the experiment compared to Pre-AD THP, Post-AD THP and Post-AD THP + Com. The gene abundance and the respiration kinetics suggested successive growth/death by different fractions of the organisms introduced with the compost and the wastewater. To inspect this further, 16S rRNA amplicons were sequenced and analyzed with bioinformatic tools.
Diversity
Bioinformatic analysis of 16S rRNA gene amplicons showed differences in both alpha and beta diversity between different biosolids (Fig. 6). Raw sequence reads were submitted to NCBI’s Sequence Read Archive (BioProject Accession ID PRJNA574619).
Microbial diversity (at OTU level) in the biosolids with and without recontamination with wastewater, shown as closed and open symbols, respectively. The diversity is expressed through Community richness and the Shannon index (a) and PCoA using the unweighted UniFrac matrix (b). The data is not TSS normalized for the alpha diversity matrices (a) but rarefied to the sample with lowest sequencing depth (55,335 reads). Additional diversity plots are provided in the Supplementary Material, Figs. S13–S19
All tested alpha-diversity matrixes (Fig. 6a; Supplementary Material, Fig. S13) indicate that compost amendment increased the microbial diversity of Post-AD THP biosolids. Accordingly, compost possessed the highest microbial diversity amongst all samples in the current experiment. It is therefore reasonable to suggest that the addition of compost overall enriches the community in Post-AD THP biosolids, thus providing an environment where E. coli is rapidly outcompeted. This complies with the lower E. coli density observed by the viable count and ddPCR results (Fig. 2). Furthermore, the multivariate analysis (Fig. 6b; Supplementary Material, Fig. S19) used to assess the community composition, demonstrated that TAD, Pre-AD THP and Post-AD THP + Com (1) had profoundly different compositions, (2) their community compositions were stable throughout the incubation, and (3) they were not much affected by recontamination with wastewater. In the sterile Post-AD THP biosolids, however, the recontamination resulted in a conspicuous temporal change in community composition (Fig. 6b; dark blue, filled symbols), suggesting successions throughout the incubation, or selective growth of subpopulations in the wastewater, reaching significant numbers at different times depending on their initial numbers and/or growth rate. This supports our interpretation of the conspicuous respiration kinetics in these biosolids: the bursts (spikes) of respiration plausibly reflect activity (hence growth) of different fractions of the bacterial community of the wastewater, thus causing major shifts in community composition. The suppression of pathogens by establishing a diverse community has an interesting parallel in aquaculture, where management that sustains a stable “K-selected” microbial community secures better survival of fish larvae than management that involves frequent disinfection [31].
Overall, these results suggest that the biosolids from Post-AD THP treatment benefit from addition of compost, providing a more robust and competitive microbial community that effectively minimize the risk for pathogen growth. It may also stabilize the biosolids by mineralizing a larger fraction of available organic material than could be achieved by contamination with wastewater alone.
Conclusion
This study presents novel insight into E. coli recontamination of various biosolids and the addition of a harmless bacteria to suppress growth of E. coli in sterilized post-anaerobic digestion thermal hydrolysis (Post-AD THP) biosolids. The pathogen concentration in a sterile Post-AD THP biosolids can meet regulatory requirements within 2 weeks of storage after recontamination, while Pre-AD THP and TAD biosolids meet the requirements 3 days after recontamination. However, pathogen growth in Post-AD THP biosolids can be suppressed by inoculation with compost to meet regulatory requirements with less storage time. Interestingly, E. coli abundance after recontamination correlated with soluble COD concentration in the biosolids. The results provide valuable input for future biosolids regulations, valorization and management.
Data availability
The sequence reads from this article have been submitted to the NCBI Sequence Read Archive under the BioProject Accession ID PRJNA574619.
References
USEPA, J.: Environmental regulations and technology: control of pathogens and vector attraction in sewage sludge. United States Environment Protection Agency, Cincinnati, OH (2003)
Fane, S., Vale, P., Bajón-Fernández, Y., Cartmell, E., Nocker, A., Harris, J., Tyrrel, S.: Influence of innate sludge factors and ambient environmental parameters in biosolids storage on indicator bacteria survival: a review. Waste Biomass Valoriz. 1–10 (2019)
Higgins, M.J., Chen, Y.C., Murthy, S.N., Hendrickson, D., Farrel, J., Schafer, P.: Reactivation and growth of non-culturable indicator bacteria in anaerobically digested biosolids after centrifuge dewatering. Water Res. 41(3), 665–673 (2007). https://doi.org/10.1016/j.watres.2006.09.017
Iranpour, R., Cox, H., Kearney, R., Clark, J., Pincince, A., Daigger, G.: Regulations for biosolids land application in US and European Union. J. Residuals Sci. Tech. 1(4), 209–222 (2004)
Landbruks- og matdepartementet: Forskrift om organisk gjødsel. Legislation on organic fertilizer. In: matdepartementet, L.-o. (ed) (2003)
Fane, S.: Control of E. coli in biosolids. PhD thesis, Cranfield University, UK (2016)
Chen, Y.C., Higgins, M.J., Beightol, S.M., Murthy, S.N., Toffey, W.E.: Anaerobically digested biosolids odor generation and pathogen indicator regrowth after dewatering. Water Res. 45(8), 2616–2626 (2011). https://doi.org/10.1016/j.watres.2011.02.014
Sidhu, J.P., Toze, S.G.: Human pathogens and their indicators in biosolids: a literature review. Environ. Int. 35(1), 187–201 (2009)
Chen, Y.-C., Higgins, M.J., Maas, N.A., Murthy, S.N.: DNA extraction and Escherichia coli quantification of anaerobically digested biosolids using the competitive touchdown PCR method. Water Res. 40(16), 3037–3044 (2006)
Hussong, D., Burge, W.D., Enkiri, N.K.: Occurrence, growth, and suppression of salmonellae in composted sewage sludge. Appl. Environ. Microb. 50(4), 887–893 (1985)
Ward, A., Stensel, H.D., Ferguson, J.F., Ma, G., Hummel, S.: Preventing growth of pathogens in pasteurized digester solids. Water Environ. Res. 71(2), 176–182 (1999)
Sidhu, J., Gibbs, R., Ho, G., Unkovich, I.: The role of indigenous microorganisms in suppression of Salmonella regrowth in composted biosolids. Water Res. 35(4), 913–920 (2001)
Murthy, S., Peot, C., Bailey, W., Higgins, M., Beightol, S., Chen, Y., Novak, J., Wilson, C.: Biosolids quality parameters important for process selection at Blue Plains AWTP: odors and indicators. In: Workshop at WEF Residuals and Biosolids Conference, Sacramento, California (2011)
Shana, A., Fountain, P., Mills, N.: SAS only THP with series digestion—more options for energy recovery. In: 18th European biosolids and organic resources conference and exhibition, Manchester, UK (2013)
Fane, S., Nocker, A., Vale, P., Casado, M.R., Cartmell, E., Harris, J., Fernández, Y.B., Tyrrel, S.: Characterisation and control of the biosolids storage environment: implications for E. coli dynamics. Sci. Total Environ. 752, 141705 (2021). https://doi.org/10.1016/j.scitotenv.2020.141705
Barber, W.P.F.: Thermal hydrolysis for sewage treatment: a critical review. Water Res. 104, 53–71 (2016). https://doi.org/10.1016/j.watres.2016.07.069
Svensson, K., Kjorlaug, O., Higgins, M.J., Linjordet, R., Horn, S.J.: Post-anaerobic digestion thermal hydrolysis of sewage sludge and food waste: effect on methane yields, dewaterability and solids reduction. Water Res. 132, 158–166 (2018). https://doi.org/10.1016/j.watres.2018.01.008
Svennevik, O.K., Solheim, O.E., Beck, G., Sørland, G.H., Jonassen, K.R., Rus, E., Westereng, B., Horn, S.J., Higgins, M.J., Nilsen, P.J.: Effects of post anaerobic digestion thermal hydrolysis on dewaterability and moisture distribution in digestates. Water Sci. Technol. 80(7), 1338–1346 (2019)
Molstad, L., Dörsch, P., Bakken, L.R.: Improved robotized incubation system for gas kinetics in batch cultures. In Technical Report (2016)
Dilly, O.: Microbial respiratory quotient during basal metabolism and after glucose amendment in soils and litter. Soil Biol. Biochem. 33(1), 117–127 (2001)
Yu, Y., Lee, C., Kim, J., Hwang, S.: Group-specific primer and probe sets to detect methanogenic communities using quantitative real-time polymerase chain reaction. Biotechnol. Bioeng. 89(6), 670–679 (2005)
Silkie, S.S., Tolcher, M.P., Nelson, K.L.: Reagent decontamination to eliminate false-positives in Escherichia coli qPCR. J. Microbiol. Methods 72(3), 275–282 (2008). https://doi.org/10.1016/j.mimet.2007.12.011
Caporaso, J.G., Kuczynski, J., Stombaugh, J., Bittinger, K., Bushman, F.D., Costello, E.K., Fierer, N., Pena, A.G., Goodrich, J.K., Gordon, J.I.: QIIME allows analysis of high-throughput community sequencing data. Nat. Methods 7(5), 335 (2010)
Quast, C., Pruesse, E., Yilmaz, P., Gerken, J., Schweer, T., Yarza, P., Peplies, J., Glöckner, F.O.: The SILVA ribosomal RNA gene database project: improved data processing and web-based tools. Nucleic Acids Res. 41(D1), D590–D596 (2012)
Zakrzewski, M., Proietti, C., Ellis, J.J., Hasan, S., Brion, M.-J., Berger, B., Krause, L.: Calypso: a user-friendly web-server for mining and visualizing microbiome–environment interactions. Bioinformatics 33(5), 782–783 (2016)
Kirkegaard, R.H., McIlroy, S.J., Kristensen, J.M., Nierychlo, M., Karst, S.M., Dueholm, M.S., Albertsen, M., Nielsen, P.H.: The impact of immigration on microbial community composition in full-scale anaerobic digesters. Sci. Rep. 7(1), 9343 (2017)
Vermeersch, L., Perez-Samper, G., Cerulus, B., Jariani, A., Gallone, B., Voordeckers, K., Steensels, J., Verstrepen, K.J.: On the duration of the microbial lag phase. Curr. Genet. 65(3), 721–727 (2019)
Clarholm, M.: Protozoan grazing of bacteria in soil—impact and importance. Microb. Ecol. 7(4), 343–350 (1981)
Pauli, W., Jax, K., Berger, S.: Protozoa in wastewater treatment: function and importance. In: Biodegradation and Persistence. pp. 203–252. Springer, New York (2001)
Bergaust, L., Mao, Y., Bakken, L.R., Frostegård, Å.: Denitrification response patterns during the transition to anoxic respiration and posttranscriptional effects of suboptimal pH on nitrogen oxide reductase in Paracoccus denitrificans. Appl. Environ. Microbiol. 76(19), 6387–6396 (2010)
Vadstein, O., Attramadal, K.J., Bakke, I., Olsen, Y.: K-selection as microbial community management strategy: a method for improved viability of larvae in aquaculture. Front. Microbiol. 9, 2730 (2018)
Acknowledgements
This work was financially supported by the Research Council of Norway (Grants no. 258749 and 260868). Biogas plants providing samples and support during this study are greatly acknowledged for their efforts. Erik Norgaard at Høst is acknowledged for supplying the compost.
Author information
Authors and Affiliations
Corresponding author
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Svennevik, O.K., Jonassen, K.R., Svensson, K. et al. Protecting Thermally Hydrolyzed Biosolids from Pathogenic Bacterial Growth by Addition of Compost. Waste Biomass Valor 12, 4263–4275 (2021). https://doi.org/10.1007/s12649-020-01300-1
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12649-020-01300-1