Abstract
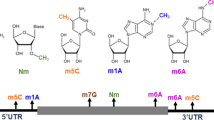
Post-transcriptional chemical modification of RNA is rapidly emerging as a key player in regulating gene expression and has propelled the development of ‘epitranscriptomics’ or ‘RNA epigenetics’ as a frontier area of research. Several RNA modifications are known to decorate RNAs and impact its structure and function. One such recently discovered modification is acetylation of RNA i.e. N4-acetylcytidine (ac4C) chemical modification. N4-acetylcytidine is an ancient and evolutionarily conserved modification, which maps to a wide spectrum of RNAs from archaea bacteria to humans. This modification results in a variety of functional outcomes which impact normal development and disease. In this review, we summarize the recent progress, emerging methods, biological implications and the future challenges for ac4C modification.

Similar content being viewed by others
References
Roundtree IA, Evans ME, Pan T, He C (2017) Dynamic RNA modifications in gene expression regulation. Cell 169(7):1187–1200. https://doi.org/10.1016/j.cell.2017.05.045
Zhao BS, Roundtree IA, He C (2017) Post-transcriptional gene regulation by mRNA modifications. Nat Rev Mol Cell Biol 18(1):31–42. https://doi.org/10.1038/nrm.2016.132
Nachtergaele S, He C (2018) Chemical modifications in the life of an mRNA transcript. Annu Rev Genet 52:349–372. https://doi.org/10.1146/annurev-genet-120417-031522
Karthiya R, Khandelia P (2020) m6A RNA methylation: ramifications for gene expression and human health. Mol Biotechnol 62(10):467–484. https://doi.org/10.1007/s12033-020-00269-5
Zhang C, Jia G (2018) Reversible RNA modification N(1)-methyladenosine (m(1)A) in mRNA and tRNA. Genomics Proteomics Bioinform 16(3):155–161. https://doi.org/10.1016/j.gpb.2018.03.003
Dominissini D, Moshitch-Moshkovitz S, Schwartz S, Salmon-Divon M, Ungar L, Osenberg S, Cesarkas K, Jacob-Hirsch J, Amariglio N, Kupiec M, Sorek R, Rechavi G (2012) Topology of the human and mouse m6A RNA methylomes revealed by m6A-seq. Nature 485(7397):201–206. https://doi.org/10.1038/nature11112
Meyer KD, Saletore Y, Zumbo P, Elemento O, Mason CE, Jaffrey SR (2012) Comprehensive analysis of mRNA methylation reveals enrichment in 3’ UTRs and near stop codons. Cell 149(7):1635–1646. https://doi.org/10.1016/j.cell.2012.05.003
Meyer KD, Jaffrey SR (2017) Rethinking m(6)A readers, writers, and erasers. Annu Rev Cell Dev Biol 33:319–342. https://doi.org/10.1146/annurev-cellbio-100616-060758
Shi H, Wei J, He C (2019) Where, when, and how: context-dependent functions of RNA methylation writers, readers, and erasers. Mol Cell 74(4):640–650. https://doi.org/10.1016/j.molcel.2019.04.025
Dominissini D, Nachtergaele S, Moshitch-Moshkovitz S, Peer E, Kol N, Ben-Haim MS, Dai Q, Di Segni A, Salmon-Divon M, Clark WC, Zheng G, Pan T, Solomon O, Eyal E, Hershkovitz V, Han D, Dore LC, Amariglio N, Rechavi G, He C (2016) The dynamic N(1)-methyladenosine methylome in eukaryotic messenger RNA. Nature 530(7591):441–446. https://doi.org/10.1038/nature16998
Arango D, Sturgill D, Alhusaini N, Dillman AA, Sweet TJ, Hanson G, Hosogane M, Sinclair WR, Nanan KK, Mandler MD, Fox SD, Zengeya TT, Andresson T, Meier JL, Coller J, Oberdoerffer S (2018) Acetylation of cytidine in mRNA promotes translation efficiency. Cell 175(7):1872–1886. https://doi.org/10.1016/j.cell.2018.10.030
Thomas JM, Briney CA, Nance KD, Lopez JE, Thorpe AL, Fox SD, Bortolin-Cavaille ML, Sas-Chen A, Arango D, Oberdoerffer S, Cavaille J, Andresson T, Meier JL (2018) A chemical signature for cytidine acetylation in RNA. J Am Chem Soc 140(40):12667–12670. https://doi.org/10.1021/jacs.8b06636
Dominissini D, Rechavi G (2018) N(4)-acetylation of cytidine in mRNA by NAT10 regulates stability and translation. Cell 175(7):1725–1727. https://doi.org/10.1016/j.cell.2018.11.037
Oashi Z, Murao K, Yahagi T, Von Minden DL, McCloskey JA, Nishimura S (1972) Characterization of C+ located in the first position of the anticodon of Escherichia coli tRNA Met as N4-acetylcytidine. Biochim Biophys Acta 262(2):209–213
Stern L, Schulman LH (1978) The role of the minor base N4-acetylcytidine in the function of the Escherichia coli noninitiator methionine transfer RNA. J Biol Chem 253(17):6132–6139
Thomas G, Gordon J, Rogg H (1978) N4-acetylcytidine. A previously unidentified labile component of the small subunit of eukaryotic ribosomes. J Biol Chem 253(4):1101–1105
Ito S, Akamatsu Y, Noma A, Kimura S, Miyauchi K, Ikeuchi Y, Suzuki T, Suzuki T (2014) A single acetylation of 18S rRNA is essential for biogenesis of the small ribosomal subunit in Saccharomyces cerevisiae. J Biol Chem 289(38):26201–26212. https://doi.org/10.1074/jbc.M114.593996
Sharma S, Langhendries JL, Watzinger P, Kotter P, Entian KD, Lafontaine DL (2015) Yeast Kre33 and human NAT10 are conserved 18S rRNA cytosine acetyltransferases that modify tRNAs assisted by the adaptor Tan1/THUMPD1. Nucleic Acids Res 43(4):2242–2258. https://doi.org/10.1093/nar/gkv075
Ikeuchi Y, Kitahara K, Suzuki T (2008) The RNA acetyltransferase driven by ATP hydrolysis synthesizes N4-acetylcytidine of tRNA anticodon. EMBO J 27(16):2194–2203. https://doi.org/10.1038/emboj.2008.154
Ito S, Horikawa S, Suzuki T, Kawauchi H, Tanaka Y, Suzuki T, Suzuki T (2014) Human NAT10 is an ATP-dependent RNA acetyltransferase responsible for N4-acetylcytidine formation in 18 S ribosomal RNA (rRNA). J Biol Chem 289(52):35724–35730. https://doi.org/10.1074/jbc.C114.602698
Chi YH, Haller K, Peloponese JM Jr, Jeang KT (2007) Histone acetyltransferase hALP and nuclear membrane protein hsSUN1 function in de-condensation of mitotic chromosomes. J Biol Chem 282(37):27447–27458. https://doi.org/10.1074/jbc.M703098200
Kong R, Zhang L, Hu L, Peng Q, Han W, Du X, Ke Y (2011) hALP, a novel transcriptional U three protein (t-UTP), activates RNA polymerase I transcription by binding and acetylating the upstream binding factor (UBF). J Biol Chem 286(9):7139–7148. https://doi.org/10.1074/jbc.M110.173393
Fu D, Collins K (2007) Purification of human telomerase complexes identifies factors involved in telomerase biogenesis and telomere length regulation. Mol Cell 28(5):773–785. https://doi.org/10.1016/j.molcel.2007.09.023
Lv J, Liu H, Wang Q, Tang Z, Hou L, Zhang B (2003) Molecular cloning of a novel human gene encoding histone acetyltransferase-like protein involved in transcriptional activation of hTERT. Biochem Biophys Res Commun 311(2):506–513. https://doi.org/10.1016/j.bbrc.2003.09.235
Liu H, Ling Y, Gong Y, Sun Y, Hou L, Zhang B (2007) DNA damage induces N-acetyltransferase NAT10 gene expression through transcriptional activation. Mol Cell Biochem 300(1–2):249–258. https://doi.org/10.1007/s11010-006-9390-5
Shen Q, Zheng X, McNutt MA, Guang L, Sun Y, Wang J, Gong Y, Hou L, Zhang B (2009) NAT10, a nucleolar protein, localizes to the midbody and regulates cytokinesis and acetylation of microtubules. Exp Cell Res 315(10):1653–1667. https://doi.org/10.1016/j.yexcr.2009.03.007
Larrieu D, Britton S, Demir M, Rodriguez R, Jackson SP (2014) Chemical inhibition of NAT10 corrects defects of laminopathic cells. Science 344(6183):527–532. https://doi.org/10.1126/science.1252651
Liu X, Tan Y, Zhang C, Zhang Y, Zhang L, Ren P, Deng H, Luo J, Ke Y, Du X (2016) NAT10 regulates p53 activation through acetylating p53 at K120 and ubiquitinating Mdm2. EMBO Rep 17(3):349–366. https://doi.org/10.15252/embr.201540505
Zhang H, Hou W, Wang HL, Liu HJ, Jia XY, Zheng XZ, Zou YX, Li X, Hou L, McNutt MA, Zhang B (2014) GSK-3beta-regulated N-acetyltransferase 10 is involved in colorectal cancer invasion. Clin Cancer Res 20(17):4717–4729. https://doi.org/10.1158/1078-0432.CCR-13-3477
Chimnaronk S, Suzuki T, Manita T, Ikeuchi Y, Yao M, Suzuki T, Tanaka I (2009) RNA helicase module in an acetyltransferase that modifies a specific tRNA anticodon. EMBO J 28(9):1362–1373. https://doi.org/10.1038/emboj.2009.69
Tardu M, Jones JD, Kennedy RT, Lin Q, Koutmou KS (2019) Identification and quantification of modified nucleosides in Saccharomyces cerevisiae mRNAs. ACS Chem Biol 14(7):1403–1409. https://doi.org/10.1021/acschembio.9b00369
Johansson MJ, Bystrom AS (2004) The Saccharomyces cerevisiae TAN1 gene is required for N4-acetylcytidine formation in tRNA. RNA 10(4):712–719. https://doi.org/10.1261/rna.5198204
Sharma S, Yang J, van Nues R, Watzinger P, Kotter P, Lafontaine DLJ, Granneman S, Entian KD (2017) Specialized box C/D snoRNPs act as antisense guides to target RNA base acetylation. PLoS Genet 13(5):e1006804. https://doi.org/10.1371/journal.pgen.1006804
Cai S, Liu X, Zhang C, Xing B, Du X (2017) Autoacetylation of NAT10 is critical for its function in rRNA transcription activation. Biochem Biophys Res Commun 483(1):624–629. https://doi.org/10.1016/j.bbrc.2016.12.092
Sleiman S, Dragon F (2019) Recent advances on the structure and function of RNA acetyltransferase Kre33/NAT10. Cells. https://doi.org/10.3390/cells8091035
Shrimp JH, Jing Y, Gamage ST, Nelson KM, Han J, Bryson KM, Montgomery DC, Thomas JM, Nance KD, Sharma S, Fox SD, Andressen T, Sinclair WR, Wu H, Allali-Hassani A, Senisterra G, Vedadi M, Lafontaine D, Dahlin JL, Marmorstein R, Walters MA, Meier JL (2020) Remodelin is a cryptic assay interference chemotype that does not inhibit NAT10-dependent cytidine acetylation. ACS Med Chem Lett. https://doi.org/10.1021/acsmedchemlett.0c00193
Kumbhar BV, Kamble AD, Sonawane KD (2013) Conformational preferences of modified nucleoside N(4)-acetylcytidine, ac4C occur at “wobble” 34th position in the anticodon loop of tRNA. Cell Biochem Biophys 66(3):797–816. https://doi.org/10.1007/s12013-013-9525-8
Igo-Kemenes T, Zachau HG (1969) On the specificity of the reduction of transfer ribonucleic acids with sodium borohydride. Eur J Biochem 10(3):549–556. https://doi.org/10.1111/j.1432-1033.1969.tb00723.x
Jegourel D, Delepee R, Breton F, Rolland A, Vidal R, Agrofoglio LA (2008) Molecularly imprinted polymer of 5-methyluridine for solid-phase extraction of pyrimidine nucleoside cancer markers in urine. Bioorg Med Chem 16(19):8932–8939. https://doi.org/10.1016/j.bmc.2008.08.063
Sakaguchi Y, Miyauchi K, Kang BI, Suzuki T (2015) Nucleoside analysis by hydrophilic interaction liquid chromatography coupled with mass spectrometry. Methods Enzymol 560:19–28. https://doi.org/10.1016/bs.mie.2015.03.015
Liebich HM, Lehmann R, Xu G, Wahl HG, Haring HU (2000) Application of capillary electrophoresis in clinical chemistry: the clinical value of urinary modified nucleosides. J Chromatogr B 745(1):189–196. https://doi.org/10.1016/s0378-4347(00)00263-2
Taoka M, Ishikawa D, Nobe Y, Ishikawa H, Yamauchi Y, Terukina G, Nakayama H, Hirota K, Takahashi N, Isobe T (2014) RNA cytidine acetyltransferase of small-subunit ribosomal RNA: identification of acetylation sites and the responsible acetyltransferase in fission yeast Schizosaccharomyces pombe. PLoS ONE 9(11):e112156. https://doi.org/10.1371/journal.pone.0112156
Sinclair WR, Arango D, Shrimp JH, Zengeya TT, Thomas JM, Montgomery DC, Fox SD, Andresson T, Oberdoerffer S, Meier JL (2017) Profiling cytidine acetylation with specific affinity and reactivity. ACS Chem Biol 12(12):2922–2926. https://doi.org/10.1021/acschembio.7b00734
Sas-Chen A, Thomas JM, Matzov D, Taoka M, Nance KD, Nir R, Bryson KM, Shachar R, Liman GLS, Burkhart BW, Gamage ST, Nobe Y, Briney CA, Levy MJ, Fuchs RT, Robb GB, Hartmann J, Sharma S, Lin Q, Florens L, Washburn MP, Isobe T, Santangelo TJ, Shalev-Benami M, Meier JL, Schwartz S (2020) Dynamic RNA acetylation revealed by quantitative cross-evolutionary mapping. Nature 583(7817):638–643. https://doi.org/10.1038/s41586-020-2418-2
Grozhik AV, Olarerin-George AO, Sindelar M, Li X, Gross SS, Jaffrey SR (2019) Antibody cross-reactivity accounts for widespread appearance of m(1)A in 5’UTRs. Nat Commun 10(1):5126. https://doi.org/10.1038/s41467-019-13146-w
Yang J, Sharma S, Watzinger P, Hartmann JD, Kotter P, Entian KD (2016) Mapping of complete set of ribose and base modifications of yeast rRNA by RP-HPLC and Mung bean nuclease assay. PLoS ONE 11(12):e0168873. https://doi.org/10.1371/journal.pone.0168873
Zhou Y, Zeng P, Li YH, Zhang Z, Cui Q (2016) SRAMP: prediction of mammalian N6-methyladenosine (m6A) sites based on sequence-derived features. Nucleic Acids Res 44(10):e91. https://doi.org/10.1093/nar/gkw104
Zhao W, Zhou Y, Cui Q, Zhou Y (2019) PACES: prediction of N4-acetylcytidine (ac4C) modification sites in mRNA. Sci Rep 9(1):11112. https://doi.org/10.1038/s41598-019-47594-7
Parsons CL, Shaw T, Berecz Z, Su Y, Zupkas P, Argade S (2014) Role of urinary cations in the aetiology of bladder symptoms and interstitial cystitis. BJU Int 114(2):286–293. https://doi.org/10.1111/bju.12603
Laguna TA, Reilly CS, Williams CB, Welchlin C, Wendt CH (2015) Metabolomics analysis identifies novel plasma biomarkers of cystic fibrosis pulmonary exacerbation. Pediatr Pulmonol 50(9):869–877. https://doi.org/10.1002/ppul.23225
Duan J, Zhang Q, Hu X, Lu D, Yu W, Bai H (2019) N(4)-acetylcytidine is required for sustained NLRP3 inflammasome activation via HMGB1 pathway in microglia. Cell Signal 58:44–52. https://doi.org/10.1016/j.cellsig.2019.03.007
Furman D, Chang J, Lartigue L, Bolen CR, Haddad F, Gaudilliere B, Ganio EA, Fragiadakis GK, Spitzer MH, Douchet I, Daburon S, Moreau JF, Nolan GP, Blanco P, Dechanet-Merville J, Dekker CL, Jojic V, Kuo CJ, Davis MM, Faustin B (2017) Expression of specific inflammasome gene modules stratifies older individuals into two extreme clinical and immunological states. Nat Med 23(2):174–184. https://doi.org/10.1038/nm.4267
Niwa T, Takeda N, Yoshizumi H (1998) RNA metabolism in uremic patients: accumulation of modified ribonucleosides in uremic serum. Techincal note. Kidney Int 53(6):1801–1806. https://doi.org/10.1046/j.1523-1755.1998.00944.x
Law KP, Han TL, Mao X, Zhang H (2017) Tryptophan and purine metabolites are consistently upregulated in the urinary metabolome of patients diagnosed with gestational diabetes mellitus throughout pregnancy: a longitudinal metabolomics study of Chinese pregnant women part 2. Clin Chim Acta 468:126–139. https://doi.org/10.1016/j.cca.2017.02.018
Bhargava P, Fitzgerald KC, Venkata SLV, Smith MD, Kornberg MD, Mowry EM, Haughey NJ, Calabresi PA (2019) Dimethyl fumarate treatment induces lipid metabolism alterations that are linked to immunological changes. Ann Clin Transl Neurol 6(1):33–45. https://doi.org/10.1002/acn3.676
Borek E, Sharma OK, Buschman FL, Cohn DL, Penley KA, Judson FN, Dobozin BS, Horsburgh CR Jr, Kirkpatrick CH (1986) Altered excretion of modified nucleosides and beta-aminoisobutyric acid in subjects with acquired immunodeficiency syndrome or at risk for acquired immunodeficiency syndrome. Cancer Res 46(5):2557–2561
Szymanska E, Markuszewski MJ, Markuszewski M, Kaliszan R (2010) Altered levels of nucleoside metabolite profiles in urogenital tract cancer measured by capillary electrophoresis. J Pharm Biomed Anal 53(5):1305–1312. https://doi.org/10.1016/j.jpba.2010.07.031
Zhang T, Wu X, Ke C, Yin M, Li Z, Fan L, Zhang W, Zhang H, Zhao F, Zhou X, Lou G, Li K (2013) Identification of potential biomarkers for ovarian cancer by urinary metabolomic profiling. J Proteome Res 12(1):505–512. https://doi.org/10.1021/pr3009572
Hua Li QQ, Shi X, He J, Guowang Xu (2019) Modified metabolites mapping by liquid chromatography-high resolution mass spectrometry using full scan/all ion fragmentation/neutral loss acquisition. J Chromatogr A 1583:80–87. https://doi.org/10.1016/j.chroma.2018.11.014
Tsai K, Jaguva Vasudevan AA, Martinez Campos C, Emery A, Swanstrom R, Cullen BR (2020) Acetylation of cytidine residues boosts HIV-1 gene expression by increasing viral RNA stability. Cell Host Microbe. https://doi.org/10.1016/j.chom.2020.05.011
Acknowledgements
P.K. acknowledges DST-SERB, Government of India for funding [Grant Number CRG/2018/000492 (to P.K.)]. Infrastructural and financial support from BITS Pilani-Hyderabad Campus, India is also acknowledged. K.R. acknowledges fellowship from BITS Pilani-Hyderabad Campus, India.
Funding
This study was funded by an extramural grant from DST-SERB, Government of India [Grant Number CRG/2018/000492 (to P.K.)].
Author information
Authors and Affiliations
Contributions
PK conceived and designed the manuscript. KR, SMW and PK wrote the manuscript. All authors have critically reviewed the manuscript and approved the final version of the manuscript.
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Ethical approval
Not required. This article does not contain any studies with human participants or animals performed by any of the authors.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Karthiya, R., Wasil, S.M. & Khandelia, P. Emerging role of N4-acetylcytidine modification of RNA in gene regulation and cellular functions. Mol Biol Rep 47, 9189–9199 (2020). https://doi.org/10.1007/s11033-020-05963-w
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11033-020-05963-w