Abstract
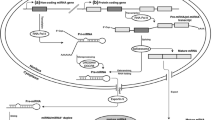
As a momentous post-transcriptional regulator, microRNAs (miRNAs) are attracting more and more attention. The classical miRNAs regulated mechanism shows it binds to the targets’ 3′UTR thus play the role in post-transcription. Meanwhile, single miRNA can target multiple genes, so those should compete to bind that miRNA. Vice versa, single gene can sponge mass of miRNAs as well. Thus the competitive endogenous RNAs (ceRNAs) hypothesis was put forward in 2011. The ceRNA hypothesis has made huge achievements, in particular in non-coding genes, which including long non-coding RNAs (lncRNAs), circle RNAs (circRNAs) and pseudogenes, even viral transcripts. It also contributed greatly to epigenetics development. However, an increasing number of controversies have occurred with applause. Based on this situation, this review introduces something in detail about the ceRNAs hypothesis achieved in lncRNAs, circRNAs, pseudogenes and viral transcripts, respectively. Meanwhile, it also covers controversy of the ceRNAs hypothesis.



Similar content being viewed by others
References
Horvitz H, Sulston J (1980) Isolation and genetic characterization of cell-lineage mutants of the nematode Caenorhabditis elegans. Genetics 96(2):435–454
Wightman B, Ha I, Ruvkun G (1993) Posttranscriptional regulation of the heterochronic gene lin-14 by lin-4 mediates temporal pattern formation in C. elegans. Cell 75(5):855–862
Lee R, Feinbaum R, Ambros V (1993) The C. elegans heterochronic gene lin-4 encodes small RNAs with antisense complementarity to lin-14. Cell 75(5):843–854. https://doi.org/10.1016/0092-8674(93)90529-y
Lee Y, Kim M, Han J, Yeom K, Lee S, Baek S, Kim V (2004) MicroRNA genes are transcribed by RNA polymerase II. EMBO J 23(20):4051–4060
Borchert G, Lanier W, Davidson B (2006) RNA polymerase III transcribes human microRNAs. Nat Struct Mol Biol 13(12):1097–1101
Lee Y, Ahn C, Han J, Choi H, Kim J, Yim J, Lee J, Provost P, Rådmark O, Kim S, Kim V (2003) The nuclear RNase III Drosha initiates microRNA processing. Nature 425(6956):415–419
Morlando M, Ballarino M, Gromak N, Pagano F, Bozzoni I, Proudfoot N (2008) Primary microRNA transcripts are processed co-transcriptionally. Nat Struct Mol Biol 15(9):902–909
Han J, Lee Y, Yeom K, Kim Y, Jin H, Kim V (2004) The Drosha-DGCR8 complex in primary microRNA processing. Genes Dev 18(24):3016–3027
Denli A, Tops B, Plasterk R, Ketting R, Hannon G (2004) Processing of primary microRNAs by the Microprocessor complex. Nature 432(7014):231–235
Yi R, Qin Y, Macara I, Cullen B (2003) Exportin-5 mediates the nuclear export of pre-microRNAs and short hairpin RNAs. Genes Dev 17(24):3011–3016
Bohnsack M, Czaplinski K, Gorlich D (2004) Exportin 5 is a RanGTP-dependent dsRNA-binding protein that mediates nuclear export of pre-miRNAs. RNA 10(2):185–191
Lund E, Güttinger S, Calado A, Dahlberg J, Kutay U (2004) Nuclear export of microRNA precursors. Science 303(5654):95–98
Ketting RF, Fischer SE, Bernstein E, Sijen T, Hannon GJ, Plasterk RH (2001) Dicer functions in RNA interference and in synthesis of small RNA involved in developmental timing in C. elegans. Genes Dev 15(20):2654–2659
Bernstein E, Caudy A, Hammond S, Hannon G (2001) Role for a bidentate ribonuclease in the initiation step of RNA interference. Nature 409(6818):363–366
Hutvágner G, McLachlan J, Pasquinelli A, Bálint E, Tuschl T, Zamore P (2001) A cellular function for the RNA-interference enzyme Dicer in the maturation of the let-7 small temporal RNA. Science 293(5531):834–838
He L, He X, Lim L, de Stanchina E, Xuan Z, Liang Y, Xue W, Zender L, Magnus J, Ridzon D, Jackson A, Linsley P, Chen C, Lowe S, Cleary M, Hannon G (2007) A microRNA component of the p53 tumour suppressor network. Nature 447(7148):1130–1134. https://doi.org/10.1038/nature05939
O'Donnell K, Wentzel E, Zeller K, Dang C, Mendell J (2005) c-Myc-regulated microRNAs modulate E2F1 expression. Nature 435(7043):839–843
Pradhan M, Prasad N, Palakal M (2012) A systems biology approach to the global analysis of transcription factors in colorectal cancer. BMC cancer 12:331
Shaulian E, Karin M (2002) AP-1 as a regulator of cell life and death. Nat Cell Biol 4(5):E131–136
Eferl R, Wagner E (2003) AP-1: a double-edged sword in tumorigenesis. Nat Rev Cancer 3(11):859–868. https://doi.org/10.1038/nrc1209
Hammond S (2015) An overview of microRNAs. Adv Drug Deliv Rev 87:3–14. https://doi.org/10.1016/j.addr.2015.05.001
Jr-Shiuan Y, Phillips MD, Doron B, Ping M, Andrea V, Siepel AC, Chen KC, Lai EC (2011) Widespread regulatory activity of vertebrate microRNA* species. RNA 17(2):312–326
Ohanian M, Humphreys D, Anderson E, Preiss T, Fatkin D (2013) A heterozygous variant in the human cardiac miR-133 gene, MIR133A2, alters miRNA duplex processing and strand abundance. BMC Genet 14:18
Salmena L, Poliseno L, Tay Y, Kats L, Pandolfi PP (2011) ceRNA hypothesis: the rosetta stone of a hidden RNA language? Cell 146(3):353–358
Tay Y, Kats L, Salmena L, Weiss D, Tan SM, Ala U, Karreth F, Poliseno L, Provero P, Dicunto F (2011) Coding-independent regulation of the tumor suppressor PTEN by competing endogenous mRNAs. Cell 147(2):344–357
Ingolia NT, Lareau LF, Weissman JS (2011) Ribosome profiling of mouse embryonic stem cells reveals the complexity and dynamics of mammalian proteomes. Cell 147(4):789–802
Mitchell G, Pamela R, Ingolia NT, Weissman JS, Lander ES (2013) Ribosome profiling provides evidence that large noncoding RNAs do not encode proteins. Cell 154(1):240–251
Sun M, Kraus WL (2015) From discovery to function: the expanding roles of long noncoding RNAs in physiology and disease. Endocr Rev 36(1):25–64
Jianchi F, Chunming B, Clark BS, Rina M, Palak S, Kohtz JD (2006) The Evf-2 noncoding RNA is transcribed from the Dlx-5/6 ultraconserved region and functions as a Dlx-2 transcriptional coactivator. Genes Dev 20(11):1470–1484
Igor M, Aroul R, Ana SB, Natalie C, Alexandre A (2007) Repression of the human dihydrofolate reductase gene by a non-coding interfering transcript. Nature 445(7128):666–670
Zhao XY, Li S, Wang GX, Yu Q, Lin J (2014) A long noncoding RNA transcriptional regulatory circuit drives thermogenic adipocyte differentiation. Mol Cell 55(3):372–382
Tichon A, Gil N, Lubelsky Y, Havkin ST, Lemze D, Itzkovitz S, Stern-Ginossar N, Ulitsky I (2016) A conserved abundant cytoplasmic long noncoding RNA modulates repression by Pumilio proteins in human cells. Nat Commun 7:12209
Kraus TFJ, Haider M, Spanner J, Steinmaurer M, Dietinger V, Kretzschmar HA (2016) Altered long noncoding rna expression precedes the course of Parkinson’s disease—a preliminary report. Mol Neurobiol 54(4):2869–2877
Sallam T, Sandhu J, Tontonoz P (2018) Long noncoding RNA discovery in cardiovascular disease: decoding form to function. Circ Res 122(1):155
Kenji T, Irene Y, Hiroaki H, Tushar P (2014) Long noncoding RNA in liver diseases. Hepatology 60(2):744–753
Martens-Uzunova ES, Böttcher R, Croce CM, Jenster G, Visakorpi T, Calin GA (2014) Long noncoding RNA in prostate, bladder, and kidney cancer. Eur Urol 65(6):1140–1151
Lin C, Yang L (2018) Long noncoding RNA in cancer: wiring signaling circuitry. Trends Cell Biol 28(4):287–301. https://doi.org/10.1016/j.tcb.2017.11.008
Wei S, Du M, Jiang Z, Hausman G, Zhang L, Dodson M (2016) Long noncoding RNAs in regulating adipogenesis: new RNAs shed lights on obesity. Cell Mol Life Sci 73(10):1–9
Kramer NJ, Gitler AD (2017) Raise the roof: boosting the efficacy of a spinal muscular atrophy therapy. Neuron 93(1):3
Jan N, Tore B, Barbara C (2007) Unexpected evidence for active brown adipose tissue in adult humans. Am J Physiol 293(2):444–452
van Lans AA, Vosselman MJ, Hanssen MJW, Brans B, Lichtenbelt WDVM (2016) Supraclavicular skin temperature and BAT activity in lean healthy adults. J Physiol Sci 66(1):77–83
Alvarez-Dominguez J, Bai Z, Xu D, Yuan B, Lo K, Yoon M, Lim Y, Knoll M, Slavov N, Chen S, Peng C, Lodish H, Sun L (2015) De novo reconstruction of adipose tissue transcriptomes reveals long non-coding RNA regulators of brown adipocyte development. Cell Metab 21(5):764–776. https://doi.org/10.1016/j.cmet.2015.04.003
Cai R, Sun Y, Qimuge N, Wang G, Wang Y, Chu G, Yu T, Yang G, Pang W (2018) Adiponectin AS lncRNA inhibits adipogenesis by transferring from nucleus to cytoplasm and attenuating Adiponectin mRNA translation. Biochem Biophys Acta 4:420
Li M, Sun X, Cai H, Sun Y, Plath M, Li C, Lan X, Lei C, Lin F, Bai Y (2016) Long non-coding RNA ADNCR suppresses adipogenic differentiation by targeting miR-204. Biochem Biophys Acta 7:871–882
Liu W, Ma C, Yang B, Yin C, Zhang B, Xiao Y (2017) LncRNA Gm15290 sponges miR-27b to promote PPARÎ3-induced fat deposition and contribute to body weight gain in mice. Biochem Biophys Res Commun 493(3):1168
Li H, Yang J, Jiang R, Wei X, Song C, Huang Y, Lan X, Lei C, Ma Y, Hu L (2018) Long non-coding RNA profiling reveals an abundant MDNCR that promotes differentiation of myoblasts by sponging miR-133a. Mol Ther Nucleic Acids 12:610–625
Zhu M, Liu J, Xiao J, Yang L, Cai M, Shen H, Chen X, Ma Y, Hu S, Wang Z (2017) Lnc-mg is a long non-coding RNA that promotes myogenesis. Nat Commun 8:14718
Dey BK, Karl P, Anindya D (2014) The H19 long noncoding RNA gives rise to microRNAs miR-675-3p and miR-675-5p to promote skeletal muscle differentiation and regeneration. Genes Dev 28(5):491–501
Liang WC, Fu WM, Wang YB, Sun YX, Xu LL, Wong CW, Chan KM, Li G, Waye MY, Zhang JF (2016) H19 activates Wnt signaling and promotes osteoblast differentiation by functioning as a competing endogenous RNA. Sci Rep 6:20121
Li K, Wu Y, Yang H, Hong P, Fang X, Hu Y (2019) H19/miR-30a/C8orf4 axis modulates the adipogenic differentiation process in human adipose tissue-derived mesenchymal stem cells. J Cell Physiol 234(11):20925–20934. https://doi.org/10.1002/jcp.28697
Cheng X, Li L, Shi G, Chen L, Fang C, Li M, Li C (2020) MEG3 promotes differentiation of porcine satellite cells by sponging miR-423–5p to relieve inhibiting effect on SRF. Cells 9(2):449. https://doi.org/10.3390/cells9020449
Jeck WR, Sorrentino JA, Wang K, Slevin MK, Burd CE, Liu J, Marzluff WF, Sharpless NE (2013) Circular RNAs are abundant, conserved, and associated with ALU repeats. RNA 19(2):141–157
Gao Y, Wang J, Zheng Y, Zhang J, Chen S, Zhao F (2016) Comprehensive identification of internal structure and alternative splicing events in circular RNAs. Nat Commun 7:12060
Zhang XO, Dong R, Zhang Y, Zhang JL, Luo Z, Zhang J, Chen LL, Yang L (2016) Diverse alternative back-splicing and alternative splicing landscape of circular RNAs. Genome Res 26(9):1277–1287
Hall I, Climent M, Quintavalle M, Farina F, Schorn T, Zani S, Carullo P, Kunderfranco P, Civilini E, Condorelli G, Elia L (2019) Circ_Lrp6, a circular RNA enriched in vascular smooth muscle cells, acts as a sponge regulating miRNA-145 function. Circ Res 124(4):498–510. https://doi.org/10.1161/circresaha.118.314240
Zhang XO, Wang HB, Zhang Y, Lu X, Chen LL, Yang L (2014) Complementary sequence-mediated exon circularization. Cell 159(1):134–147
Xintian Y, Irena V, Ana B, Tristan W, Irina E, Georgi T, Güney A, Mantian W, Caspar G, Claudia Q (2015) Neural circular RNAs are derived from synaptic genes and regulated by development and plasticity. Nat Neurosci 18(4):603–610
Dong R, Zhang XO, Zhang Y, Ma XK, Chen LL, Yang L (2016) CircRNA-derived pseudogenes. Cell Res 26(6):747–750
Agnieszka RW, Christin S, Petar GA, Marvin J, Natalia P, Sebastian G, Mor H, Mikaela B, Osnat B, Reut AF (2015) Circular RNAs in the mammalian brain are highly abundant, conserved, and dynamically expressed. Mol Cell 58(5):870–885
Chen YG, Kim MV, Chen X, Batista PJ, Aoyama S, Wilusz JE, Iwasaki A, Chang HY (2017) Sensing self and foreign circular RNAs by intron identity. Mol Cell 67(2):S1097276517303623
Hansen TB, Jensen TI, Clausen BH, Bramsen JB, Bente F, Damgaard CK, Kjems J (2013) Natural RNA circles function as efficient microRNA sponges. Nature 495(7441):384–388
Li X, Zheng Y, Zheng Y, Huang Y, Zhang Y, Jia L, Li W (2018) Circular RNA CDR1as regulates osteoblastic differentiation of periodontal ligament stem cells via the miR-7/GDF5/SMAD and p38 MAPK signaling pathway. Stem Cell Res Ther 9(1):232
Wang K, Gan TY, Li N, Liu CY, Zhou LY, Gao JN, Chen C, Yan KW, Ponnusamy M, Zhang YH (2017) Circular RNA mediates cardiomyocyte death via miRNA-dependent upregulation of MTP18 expression. Cell Death Differ 24(6):1111–1120
Yang C, Yuan W, Yang X, Li P, Wang J, Han J, Tao J, Li P, Yang H, Lv Q (2018) Circular RNA circ-ITCH inhibits bladder cancer progression by sponging miR-17/miR-224 and regulating p21. PTEN Expr Mol Cancer 17(1):19
Zhang Z, Zhang T, Feng R, Huang H, Xia T, Sun C (2018) circARF3 alleviates mitophagy-mediated inflammation by targeting miR-103/TRAF3 in mouse adipose tissue. Mol Ther-Nucleic Aicds 14(12):192–203
Shan K, Liu C, Liu BH, Chen X, Dong R, Liu X, Zhang YY, Liu B, Zhang SJ, Wang JJ (2017) Circular non-coding RNA HIPK3 mediates retinal vascular dysfunction in diabetes mellitus. Circulation 136(17):1629
Wang X, Cao X, Dong D, Shen X, Cheng J, Jiang R, Yang Z, Peng S, Huang Y, Lan X, Elnour I, Lei C, Chen H (2019) Circular RNA TTN acts as a miR-432 sponge to facilitate proliferation and differentiation of myoblasts via the IGF2/PI3K/AKT signaling pathway. Mol Ther Nucleic acids 18:966–980. https://doi.org/10.1016/j.omtn.2019.10.019
Shen X, Zhang X, Ru W, Huang Y, Lan X, Lei C, Chen H (2020) circINSR promotes proliferation and reduces apoptosis of embryonic myoblasts by sponging miR-34a. Mol Ther Nucleic Acids 19:986–999. https://doi.org/10.1016/j.omtn.2019.12.032
Yue B, Wang J, Ru W, Wu J, Cao X, Yang H, Huang Y, Lan X, Lei C, Huang B, Chen H (2020) The circular RNA circHUWE1 sponges the miR-29b-AKT3 axis to regulate myoblast development. Mol Ther Nucleic acids 19:1086–1097. https://doi.org/10.1016/j.omtn.2019.12.039
Liu Y, Liu H, Li Y, Mao R, Yang H, Zhang Y, Zhang Y, Guo P, Zhan D, Zhang T (2020) Circular RNA SAMD4A controls adipogenesis in obesity through the miR-138-5p/EZH2 axis. Theranostics 10(10):4705–4719. https://doi.org/10.7150/thno.42417
Goodhead I, Darby AC (2015) Taking the pseudo out of pseudogenes. Curr Opin Microbiol 23:102–109
Poliseno L (2012) Pseudogenes: newly discovered players in human cancer. Sci Signal 5(242):re5. https://doi.org/10.1126/scisignal.2002858
Karreth F, Reschke M, Ruocco A, Ng C, Chapuy B, Léopold V, Sjoberg M, Keane T, Verma A, Ala U (2015) The BRAF pseudogene functions as a competitive endogenous RNA and induces lymphoma invivo. Cell 161(2):319–332
Kalyana-Sundaram S, Kumar-Sinha C, Shankar S, Robinson D, Wu YM, Cao X, Asangani I, Kothari V, Prensner J, Lonigro R (2012) Expressed pseudogenes in the transcriptional landscape of human cancers. Cell 149(7):1622–1634
Poliseno L, Pandolfi PP (2015) PTEN ceRNA networks in human cancer. Methods 77–78:41–50
Poliseno L, Salmena L, Zhang J, Carver B, Haveman W, Pandolfi P (2010) A coding-independent function of gene and pseudogene mRNAs regulates tumour biology. Nature 465(7301):1033–1038. https://doi.org/10.1038/nature09144
Rutnam ZJ, Du WW, Yang W, Yang X, Yang BB (2014) The pseudogene TUSC2P promotes TUSC2 function by binding multiple microRNAs. Nat Commun 5(1):2914
Lv H, Tong J, Yang J, Lv S, Li W, Zhang C, Chen Z (2018) HK2P1Dysregulated pseudogene may contribute to preeclampsia as a competing endogenous RNA for hexokinase 2 by impairing decidualization. Hypertension 71(4):648–658. https://doi.org/10.1161/hypertensionaha.117.10084
Lufeng Z, Xiaoman L, Yi G, Xiaobo L, Tao X (2015) The 3'UTR of the pseudogene CYP4Z2P promotes tumor angiogenesis in breast cancer by acting as a ceRNA for CYP4Z1. Breast Cancer Res Treat 150(1):105–118
Zheng L, Li X, Gu Y, Lv X, Xi T (2020) Correction to: The 3'UTR of the pseudogene CYP4Z2P promotes tumor angiogenesis in breast cancer by acting as a ceRNA for CYP4Z1. Breast Cancer Res Treat 179(2):521–522. https://doi.org/10.1007/s10549-019-05478-4
Tian X, Song J, Zhang X, Yan M, Wang S, Wang Y, Xu L, Zhao L, Wei J, Shao C, Kong B, Liu Z (2020) MYC-regulated pseudogene HMGA1P6 promotes ovarian cancer malignancy via augmenting the oncogenic HMGA1/2. Cell Death Dis 11(3):167. https://doi.org/10.1038/s41419-020-2356-9
Wang S, Qi Y, Gao X, Qiu W, Liu Q, Guo X, Qian M, Chen Z, Zhang Z, Wang H, Xu J, Xue H, Guo X, Zhang P, Zhao R, Li G (2020) Hypoxia-induced lncRNA PDIA3P1 promotes mesenchymal transition via sponging of miR-124-3p in glioma. Cell Death Dis 11(3):168. https://doi.org/10.1038/s41419-020-2345-z
Du C, Wang H, Chen P, Chen C (2019) STAT3-induced upregulation of lncRNA DUXAP8 functions as ceRNA for miR-577 to promote the migration and invasion in colorectal cancer through the regulation of RAB14. Eur Rev Med Pharmacol Sci 23(14):6105–6118. https://doi.org/10.26355/eurrev_201907_18424
Gottwein E, Cullen B (2008) Viral and cellular microRNAs as determinants of viral pathogenesis and immunity. Cell Host Microbe 3(6):375–387
Bogerd H, Skalsky R, Kennedy E, Furuse Y, Whisnant A, Flores O, Schultz K, Putnam N, Barrows N, Sherry B, Scholle F, Garcia-Blanco M, Griffin D, Cullen B (2014) Replication of many human viruses is refractory to inhibition by endogenous cellular microRNAs. J Virol 88(14):8065–8076
Ghosh Z, Mallick B, Chakrabarti J (2009) Cellular versus viral microRNAs in host-virus interaction. Nucleic Acids Res 37(4):1035–1048. https://doi.org/10.1093/nar/gkn1004
Dunn W, Trang P, Zhong Q, Yang E, van Belle C, Liu F (2005) Human cytomegalovirus expresses novel microRNAs during productive viral infection. Cell Microbiol 7(11):1684–1695
Umbach J, Kramer M, Jurak I, Karnowski H, Coen D, Cullen B (2008) MicroRNAs expressed by herpes simplex virus 1 during latent infection regulate viral mRNAs. Nature 454(7205):780–783
Pfeffer S, Zavolan M, Grässer F, Chien M, Russo J, Ju J, John B, Enright A, Marks D, Sander C, Tuschl T (2004) Identification of virus-encoded microRNAs. Science 304(5671):734–736. https://doi.org/10.1126/science.1096781
Cai X, Lu S, Zhang Z, Gonzalez C, Damania B, Cullen B (2005) Kaposi's sarcoma-associated herpesvirus expresses an array of viral microRNAs in latently infected cells. Proc Natl Acad Sci USA 102(15):5570–5575
Pfeffer S, Sewer A, Lagos-Quintana M, Sheridan R, Sander C, Grässer F, van Dyk L, Ho C, Shuman S, Chien M, Russo J, Ju J, Randall G, Lindenbach B, Rice C, Simon V, Ho D, Zavolan M, Tuschl T (2005) Identification of microRNAs of the herpesvirus family. Nat Methods 2(4):269–276. https://doi.org/10.1038/nmeth746
Hussain M, Taft R, Asgari S (2008) An insect virus-encoded microRNA regulates viral replication. J Virol 82(18):9164–9170
Jopling C, Yi M, Lancaster A, Lemon S, Sarnow P (2005) Modulation of hepatitis C virus RNA abundance by a liver-specific MicroRNA. Science 309(5740):1577–1581
Zhou J, Yu L, Gao X, Hu J, Wang J, Dai Z, Wang J, Zhang Z, Lu S, Huang X, Wang Z, Qiu S, Wang X, Yang G, Sun H, Tang Z, Wu Y, Zhu H, Fan J (2011) Plasma microRNA panel to diagnose hepatitis B virus-related hepatocellular carcinoma. J Clin Oncol 29(36):4781–4788
Xiong Y, Zhang C, Yuan J, Zhu Y, Tan Z, Kuang X, Wang X (2015) Hepatitis C virus represses the cellular antiviral response by upregulating the expression of signal transducer and activator of transcription 3 through sponging microRNA-122. Mol Med Rep 11(3):1733–1737. https://doi.org/10.3892/mmr.2014.2897
Cazalla D, Yario T, Steitz J, Steitz J (2010) Down-regulation of a host microRNA by a Herpesvirus saimiri noncoding RNA. Science 328(5985):1563–1566
Liu N, Jiao T, Huang Y, Liu W, Li Z, Ye X (2015) Hepatitis B virus regulates apoptosis and tumorigenesis through the microRNA-15a-Smad7-transforming growth factor beta pathway. J Virol 89(5):2739–2749
Liu N, Zhang J, Jiao T, Li Z, Peng J, Cui Z, Ye X (2013) Hepatitis B virus inhibits apoptosis of hepatoma cells by sponging the MicroRNA 15a/16 cluster. J Virol 87(24):13370–13378
Hon L, Zhang Z (2007) The roles of binding site arrangement and combinatorial targeting in microRNA repression of gene expression. Genome Biol 8(8):R166
Denzler R, Agarwal V, Stefano J, Bartel D, Stoffel M (2014) Assessing the ceRNA hypothesis with quantitative measurements of miRNA and target abundance. Mol Cell 54(5):766–776
Bosson A, Zamudio J, Sharp P (2014) Endogenous miRNA and target concentrations determine susceptibility to potential ceRNA competition. Mol Cell 56(3):347–359
Ebert M, Neilson J, Sharp P (2007) MicroRNA sponges: competitive inhibitors of small RNAs in mammalian cells. Nat Methods 4(9):721–726
Jens M, Rajewsky N (2015) Competition between target sites of regulators shapes post-transcriptional gene regulation. Nat Rev Genet 16(2):113–126
Bissels U, Wild S, Tomiuk S, Holste A, Hafner M, Tuschl T, Bosio A (2009) Absolute quantification of microRNAs by using a universal reference. RNA 15(12):2375–2384
Calabrese J, Seila A, Yeo G, Sharp P (2007) RNA sequence analysis defines Dicer's role in mouse embryonic stem cells. Proc Natl Acad Sci USA 104(46):18097–18102
Mukherji S, Ebert M, Zheng G, Tsang J, Sharp P, van Oudenaarden A (2011) MicroRNAs can generate thresholds in target gene expression. Nat Genet 43(9):854–859. https://doi.org/10.1038/ng.905
Yuan Y, Liu B, Xie P, Zhang M, Li Y, Xie Z, Wang X (2015) Model-guided quantitative analysis of microRNA-mediated regulation on competing endogenous RNAs using a synthetic gene circuit. Proc Natl Acad Sci USA 112(10):3158–3163
Nielsen C, Shomron N, Sandberg R, Hornstein E, Kitzman J, Burge C (2007) Determinants of targeting by endogenous and exogenous microRNAs and siRNAs. RNA 13(11):1894–1910
Friedman R, Farh K, Burge C, Bartel D (2009) Most mammalian mRNAs are conserved targets of microRNAs. Genome Res 19(1):92–105. https://doi.org/10.1101/gr.082701.108
Brennecke J, Stark A, Russell R, Cohen S (2005) Principles of microRNA-target recognition. PLoS Biol 3(3):e85
Grimson A, Farh K, Johnston W, Garrett-Engele P, Lim L, Bartel D (2007) MicroRNA targeting specificity in mammals: determinants beyond seed pairing. Mol Cell 27(1):91–105
Sheu-Gruttadauria J, Xiao Y, Gebert L, MacRae I (2019) Beyond the seed: structural basis for supplementary microRNA targeting by human Argonaute2. EMBO J 38(13):e101153. https://doi.org/10.15252/embj.2018101153
Loinger A, Shemla Y, Simon I, Margalit H, Biham O (2012) Competition between small RNAs: a quantitative view. Biophys J 102(8):1712–1721
Min K, Zealy R, Davila S, Fomin M, Cummings J, Makowsky D, Mcdowell C, Thigpen H, Hafner M, Kwon S, Georgescu C, Wren J, Yoon J (2018) Profiling of m6A RNA modifications identified an age-associated regulation of AGO2 mRNA stability. Aging Cell 17(3):e12753. https://doi.org/10.1111/acel.12753
Helwak A, Kudla G, Dudnakova T, Tollervey D (2013) Mapping the human miRNA interactome by CLASH reveals frequent noncanonical binding. Cell 153(3):654–665
Chi S, Hannon G, Darnell R (2012) An alternative mode of microRNA target recognition. Nat Struct Mol Biol 19(3):321–327
Brümmer A, Yang Y, Chan T, Xiao X (2017) Structure-mediated modulation of mRNA abundance by A-to-I editing. Nat Commun 8(1):1255. https://doi.org/10.1038/s41467-017-01459-7
Thomson DW, Dinger ME (2016) Endogenous microRNA sponges: evidence and controversy. Nat Rev Genet 17(5):272
Kartha R, Subramanian S (2014) Competing endogenous RNAs (ceRNAs): new entrants to the intricacies of gene regulation. Front Genet 5:8. https://doi.org/10.3389/fgene.2014.00008
Lewis B, Burge C, Bartel D (2005) Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are microRNA targets. Cell 120(1):15–20
John B, Enright A, Aravin A, Tuschl T, Sander C, Marks D (2004) Human MicroRNA targets. PLoS Biol 2(11):e363
Miranda K, Huynh T, Tay Y, Ang Y, Tam W, Thomson A, Lim B, Rigoutsos I (2006) A pattern-based method for the identification of MicroRNA binding sites and their corresponding heteroduplexes. Cell 126(6):1203–1217
Krek A, Grün D, Poy M, Wolf R, Rosenberg L, Epstein E, MacMenamin P, da Piedade I, Gunsalus K, Stoffel M, Rajewsky N (2005) Combinatorial microRNA target predictions. Nat Genet 37(5):495–500. https://doi.org/10.1038/ng1536
Kertesz M, Iovino N, Unnerstall U, Gaul U, Segal E (2007) The role of site accessibility in microRNA target recognition. Nat Genet 39(10):1278–1284
Mathonnet G, Fabian M, Svitkin Y, Parsyan A, Huck L, Murata T, Biffo S, Merrick W, Darzynkiewicz E, Pillai R, Filipowicz W, Duchaine T, Sonenberg N (2007) MicroRNA inhibition of translation initiation in vitro by targeting the cap-binding complex eIF4F. Science 317(5845):1764–1767. https://doi.org/10.1126/science.1146067
Chiu H, Martínez M, Komissarova E, Llobet-Navas D, Bansal M, Paull E, Silva J, Yang X, Sumazin P, Califano A (2018) The number of titrated microRNA species dictates ceRNA regulation. Nucleic Acids Res 46(9):4354–4369. https://doi.org/10.1093/nar/gky286
Poria DK, Guha A, Nandi I, Ray PS (2016) RNA-binding protein HuR sequesters microRNA-21 to prevent translation repression of proinflammatory tumor suppressor gene programmed cell death 4. Oncogene 35(13):1703–1715
Floor S, Doudna J (2015) Get in LINE: competition for newly minted retrotransposon proteins at the ribosome. Mol Cell 60(5):712–714
Chen K, Xie S, Jin W (2019) Crucial lncRNAs associated with adipocyte differentiation from human adipose-derived stem cells based on co-expression and ceRNA network analyses. PeerJ 7:e7544. https://doi.org/10.7717/peerj.7544
Liu Y, Wang Y, He X, Zhang S, Wang K, Wu H, Chen L (2018) LncRNA TINCR/miR-31-5p/C/EBP-α feedback loop modulates the adipogenic differentiation process in human adipose tissue-derived mesenchymal stem cells. Stem cell Res 32:35–42. https://doi.org/10.1016/j.scr.2018.08.016
Li R, Li B, Shen M, Cao Y, Zhang X, Li W, Tao J, Wu W, Liu H (2020) LncRNA 2310043L19Rik inhibits differentiation and promotes proliferation of myoblast by sponging miR-125a-5p. Aging 12(7):5625–5639. https://doi.org/10.18632/aging.102905
Li Z, Cai B, Abdalla B, Zhu X, Zheng M, Han P, Nie Q, Zhang X (2019) LncIRS1 controls muscle atrophy via sponging miR-15 family to activate IGF1-PI3K/AKT pathway. J Cachexia Sarcopenia Muscle 10(2):391–410. https://doi.org/10.1002/jcsm.12374
Zhang T, Liu H, Mao R, Yang H, Zhang Y, Zhang Y, Guo P, Zhan D, Xiang B, Liu Y (2020) The lncRNA RP11–142A22.4 promotes adipogenesis by sponging miR-587 to modulate Wnt5β expression. Cell Death Dis 11(6):475. https://doi.org/10.1038/s41419-020-2550-9
Chen Y, Li K, Zhang X, Chen J, Li M, Liu L (2020) The novel long noncoding RNA lncRNA-Adi regulates adipogenesis. Stem Cells Transl Med. https://doi.org/10.1002/sctm.19-0438
Sun Y, Li Y, Wang M, Yue M, Bai L, Bian J, Hao W, Sun J, Zhang S, Liu H (2020) Increased ATR expression is induced by ATR autoantibody via two axes, Klf-5/IRF-1 and circErbB4/miR-29a-5p, to promote VSMC migration. Cell Death Dis 11(6):432. https://doi.org/10.1038/s41419-020-2643-5
Lin J, Feng X, Zhang J (2020) Circular RNA circHIPK3 modulates the proliferation of airway smooth muscle cells by miR-326/STIM1 axis. Life Sci 255:117835. https://doi.org/10.1016/j.lfs.2020.117835
Shen X, Liu Z, Cao X, He H, Han S, Chen Y, Cui C, Zhao J, Li D, Wang Y, Zhu Q, Yin H (2019) Circular RNA profiling identified an abundant circular RNA circTMTC1 that inhibits chicken skeletal muscle satellite cell differentiation by sponging miR-128-3p. Int J Biol Sci 15(10):2265–2281. https://doi.org/10.7150/ijbs.36412
Li L, Chen Y, Nie L, Ding X, Zhang X, Zhao W, Xu X, Kyei B, Dai D, Zhan S, Guo J, Zhong T, Wang L (1862) Zhang H (2019) MyoD-induced circular RNA CDR1as promotes myogenic differentiation of skeletal muscle satellite cells. Biochim Biophys Acta 8:807–821. https://doi.org/10.1016/j.bbagrm.2019.07.001
Peng S, Song C, Li H, Cao X, Ma Y, Wang X, Huang Y, Lan X, Lei C, Chaogetu B, Chen H (2019) Circular RNA SNX29 sponges miR-744 to regulate proliferation and differentiation of myoblasts by activating the Wnt5a/Ca signaling pathway. Mol Ther Nucleic Acids 16:481–493. https://doi.org/10.1016/j.omtn.2019.03.009
Lai Y, Li J, Zhong L, He X, Si X, Sun Y, Chen Y, Zhong J, Hu Y, Li B, Liao W, Liu C, Liao Y, Xiu J, Bin J (2019) The pseudogene PTENP1 regulates smooth muscle cells as a competing endogenous RNA. Clin Sci 133(13):1439–1455. https://doi.org/10.1042/cs20190156
Hao S, Ma H, Niu Z, Sun S, Zou Y, Xia H (2019) hUC-MSCs secreted exosomes inhibit the glioma cell progression through PTENP1/miR-10a-5p/PTEN pathway. Eur Rev Med Pharmacol Sci 23(22):10013–10023. https://doi.org/10.26355/eurrev_201911_19568
Wang C, Wang L, Yang T, Su S, Hu Y, Zhong D (2020) Pseudogene PTENP1 sponges miR-214 to regulate the expression of PTEN to modulate osteoclast differentiation and attenuate osteoporosis. Cytotherapy. https://doi.org/10.1016/j.jcyt.2020.04.090
Ou L, Xiang T, Hao X, Wang D, Zeng Q (2020) Reduced long non-coding RNA PTENP1 contributed to proliferation and invasion via miR-19b/MTUS1 axis in patients with cervical cancer. Eur Rev Med Pharmacol Sci 24(8):4132–4144. https://doi.org/10.26355/eurrev_202004_20993
Sharma S, Chatterjee A, Kumar P, Lal S, Kondabagil K (2020) Upregulation of miR-101 during influenza A virus infection abrogates viral life cycle by targeting mTOR pathway. Viruses 12(4):444. https://doi.org/10.3390/v12040444
Acknowledgements
This work was supported by the National Natural Science Foundation of China (Grant numbers 31872334, 31902132).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Lin, W., Liu, H., Tang, Y. et al. The development and controversy of competitive endogenous RNA hypothesis in non-coding genes. Mol Cell Biochem 476, 109–123 (2021). https://doi.org/10.1007/s11010-020-03889-2
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11010-020-03889-2