Abstract
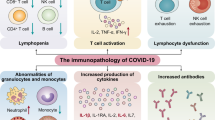
Severe acute respiratory syndrome coronavirus 2 infections can cause coronavirus disease 2019 (COVID-19), which manifests with a range of severities from mild illness to life-threatening pneumonia and multi-organ failure. Severe COVID-19 is characterized by an inflammatory signature, including high levels of inflammatory cytokines, alveolar inflammatory infiltrates and vascular microthrombi. Here we show that patients with severe COVID-19 produced a unique serologic signature, including an increased likelihood of IgG1 with afucosylated Fc glycans. This Fc modification on severe acute respiratory syndrome coronavirus 2 IgGs enhanced interactions with the activating Fcγ receptor FcγRIIIa; when incorporated into immune complexes, Fc afucosylation enhanced production of inflammatory cytokines by monocytes, including interleukin-6 and tumor necrosis factor. These results show that disease severity in COVID-19 correlates with the presence of proinflammatory IgG Fc structures, including afucosylated IgG1.
Similar content being viewed by others
Main
Antibody responses to viral infections in humans are varied and of widely divergent clinical significance. Preexisting, reactive antibodies or antibodies that are formed early during infection can bind to virus particles, forming immune complexes that can neutralize or mediate clearance of the virus. On the other hand, immune complexes can also promote inflammation and exacerbate symptoms of disease. How antibodies within immune complexes modulate infection depends, in part, on their Fc domain structure. Fc structure, in turn, dictates interactions with Fcγ receptors (FcγRs), which are expressed by a variety of cells that are activated during infection1.
Antibody isotypes IgG, IgA and IgM are a primary determinant of Fc structure and thus of activity. Initial B cell responses are characterized by the production of IgM antibodies. Production of class-switched IgA and IgG antibodies follows, with IgA playing a central role in mucosal immunity, while IgG is the dominant isotype involved in systemic antiviral immunity. IgG functions are governed by interactions between immune complexes and effector immune cells that express FcγRs, the receptors for IgG. The balance of FcγRs that are engaged by immune complexes determines the degree of the inflammatory effector cell response. Activating, low-affinity FcγRs (FcγRIIa and FcγRIIIa) transduce inflammatory signaling through immunoreceptor tyrosine-based activation motifs (ITAMs); in health, ITAM signaling is balanced by immunoreceptor tyrosine-based inhibition motif (ITIM) signaling through the inhibitory FcγRIIb2,3.
The strength of interactions between immune complexes and various FcγRs is determined by structural diversity within IgG subclasses (IgG1, IgG2, IgG3 and IgG4) and posttranslational modifications of their Fc domains4. Importantly, individuals produce distinct structural repertoires of IgG Fc domains, with some producing highly activating/proinflammatory repertoires enriched for features such as IgG1, IgG3 and/or reduced core fucosylation of the IgG1 Fc domain. Others produce IgG repertoires characterized by higher levels of IgG2 and/or sialylated Fcs that have reduced activating/inflammatory FcγR signaling potential2. It remains unknown whether any specific structural features of antibodies are produced by patients with mild or severe COVID-19.
To address this, we performed a multidimensional analysis of antibodies from adult patients with severe (hospitalized) or mild (outpatient) severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) infections, or from SARS-CoV-2 seropositive children, with a focus on antibody features that are known to augment effector functions. Overall, we found that severe COVID-19 was distinguished by increased production of IgG1 antibodies with significantly reduced Fc fucosylation (high afucosylation) and elevated IgG3 in patients who were treated in the intensive care unit (ICU), both of these antibody forms being proinflammatory in nature. Within SARS-CoV-2 immune complexes, afucosylated Fc structures triggered activation of natural killer (NK) cells and production of inflammatory cytokines by primary monocytes including interleukin (IL)-6 and tumor necrosis factor (TNF).
Results
Patients with severe COVID-19 display specific antibody signatures
A total of 43 patients hospitalized with PCR+ COVID-19, including those treated in the ICU (n = 21) or on the floor (non-ICU; n = 22), along with 18 outpatients with PCR+ COVID-19 were studied (Table 1). In addition, because children are rarely diagnosed with COVID-19 and almost never develop severe COVID-19 despite being susceptible to productive infections5,6,7,8, we reasoned that it would be informative to study SARS-CoV-2 antibodies produced by children. To this end, we screened approximately 800 remainder sera from pediatric patients in a large Northern Californian healthcare system and identified 16 that were positive for antibodies against the receptor-binding domain (RBD) of the SARS-CoV-2 spike protein; all positive samples from children were validated in a secondary screen against the full-length spike protein, as previously described9.
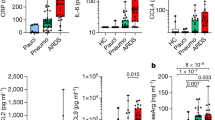
To characterize serologic correlates of disease severity, we first profiled anti-RBD immunoglobulin isotype titers in sera from patients with COVID-19 or from seropositive children. Among the study participants, patients with severe COVID-19 (ICU and floor) had significantly higher serum titers of IgM and IgA RBD-binding antibodies compared to patients with mild COVID-19 and seropositive children. The anti-RBD IgG titers were not significantly different among the groups (Fig. 1a,b).
a, Anti-RBD IgM, IgA and IgG titers in patients with COVID-19 who required treatment in the ICU (red; n = 21), hospitalization but no ICU (floor; yellow; n = 22) or treatment on an outpatient basis (light blue; n = 18), and seropositive children (peds; dark blue; n = 16). OD, optical density. b, Anti-RBD AUC for the four patient groups. Data are representative of at least two experiments performed in triplicate. Violin plots show the distribution of sample values along with median (solid lines) and upper and lower quartile (dotted lines) values. The dashed line indicates the average AUC value of the pre-COVID-19 historical samples. P values in b were calculated using one-way ANOVA with Tukey’s multiple-comparisons testing between all groups. IgM: P = 0.0079, ICU versus outpatient; P = 0.0079, ICU versus peds; P = 0.0023, floor versus outpatient; and P = 0.0023, floor versus peds. IgA: P = 0.0005, ICU versus outpatient; P = 0.0001, ICU versus peds; P = 0.0454, floor versus outpatient; and P = 0.0148, floor versus peds. *P ≤ 0.05, **P ≤ 0.01, ***P ≤ 0.001 and ****P ≤ 0.0001.
Next, we defined the determinants of anti-RBD IgG effector function in the patients with COVID-19 and seropositive children. First, the absolute abundance of various IgG subclasses was characterized by mass spectrometry. We observed that the abundance of different subclasses was similar between groups, with a small but significant increase in IgG3 produced by patients with COVID-19 who were in the ICU (Fig. 2a). Anti-RBD IgG1, the most abundant IgG subclass, was next characterized for posttranslational modifications of the Fc domain using well-established mass spectrometric methods10,11,12. The relative abundances of N-glycan modifications were characterized, including fucose, bisecting N-acetyl glucosamine (GlcNAc), galactose and sialic acid. Notably, anti-RBD IgG1 from patients with severe COVID-19 was significantly reduced in core fucosylation when compared with anti-RBD IgG1 from patients with mild COVID-19 or from seropositive children (Fig. 2b,c). We validated the ability of reduced fucosylation to separate patients with severe COVID-19 from those with mild disease using receiver operating characteristic (ROC) curve analysis (global area under the curve (AUC) value of 0.74 (95% confidence interval (CI) 0.62–0.86), P = 0.0031; Fig. 2d). Of all posttranslational modifications, IgG1 Fc fucosylation was a uniquely strong marker of severe COVID-19 using ROC curve analysis (Extended Data Fig. 1). When we analyzed the six afucosylated Fc glycoforms that were quantified, those without a bisecting N-acetyl glucosamine (F0N0) were specifically increased in severe COVID-19 (Fig. 2e).
a, The abundances of anti-RBD IgG subclasses were characterized. ICU patients had elevated levels of IgG3 compared to those treated on the floor or outpatients. P values were calculated using unpaired two-tailed t-tests (P = 0.0449, ICU versus floor; P = 0.0243, ICU versus outpatient). b, Cartoon representation of IgG1 Fc glycans and various F0 modifications. c, Anti-RBD IgG1 Fc posttranslational modifications. Patients who were hospitalized (ICU or floor) had significantly reduced Fc fucosylation (F) when compared with RBD IgGs from outpatients (P = 0.0092, ICU versus outpatient; P = 0.0011, floor versus outpatient) or from children (P = 0.0001, ICU versus peds; P ≤ 0.0001, floor versus peds). Fc galactosylation (GS0) was significantly higher and sialylation (S) significantly lower in all adult patients compared with children. GS0: P = 0.0019, ICU versus peds; P = 0.0007, floor versus peds; P = 0.0007, outpatient versus peds. S: P = 0.0097, ICU versus peds; P = 0.0045, floor versus peds; P = 0.0052, outpatient versus peds). No significant differences were observed in levels of IgG1 Fc bisection (N). d, ROC curve analysis for anti-RBD IgG1 fucosylation from all patients with COVID-19 who were hospitalized (n = 43) or had mild disease (n = 18) showed IgG1 fucose levels separated the two cohorts. AUC: 0.74 (95% CI: 0.62–0.86); P = 0.0031. e, Of the six afucosylated forms quantified, those lacking both core fucose and a bisecting N-acetyl glucosamine (F0N0) were substantially enriched in patients with severe COVID-19. Violin plots show the distribution of sample values along with median (solid lines) and upper and lower quartile (dotted lines) values. P values in c and e were calculated using one-way ANOVA with Tukey’s multiple-comparisons testing between all groups. *P ≤ 0.05, **P ≤ 0.01, ***P ≤ 0.001 and ****P ≤ 0.0001.
Overall, these data showed a unique antibody signature associated with COVID-19 severity based on multiparametric characterization of SARS-CoV-2-specific humoral responses comprising isotype titers, IgG subclasses and IgG1 Fc glycan structures. Patients with severe COVID-19 were more likely to produce significantly higher titers of anti-RBD IgM and IgA isotypes, increased IgG3 (ICU patients) and increased afucosylated (F0N0) Fc glycoforms compared to patients with mild COVID-19 (Fig. 3). The IgG3 subclass and afucosylated IgG1 modifications are features that increase Fc interactions with activating/proinflammatory FcγRs2.
Relative multidimensional antibody signatures for each group stratified by disease severity are depicted by radar plots. Each feature, isotype (IgM, IgA and IgG; blue), IgG subclass (IgG1, IgG2, IgG3 and IgG4; yellow) and percentage abundance of IgG1 Fc glycoforms (F0N0, GS0, S and N; red) is depicted as a wedge. The size of the wedge indicates the median of the features, normalized to the corresponding outpatient feature.
Fc afucosylation is stable over time in severe COVID-19
The afucosylated IgG1 Fc was the most prominent feature distinguishing severe and mild COVID-19 in our cohorts. To begin to dissect the regulation of this antibody modification, we first asked whether infection itself triggered a change in the abundance of any of the Fc glycoforms. To do this, we performed a longitudinal analysis of Fc glycoforms on RBD-reactive IgG1 from paired sera drawn at acute (T1) and later time points. Paired samples were selected from individuals who were positive for IgG at T1 and late time points were drawn 2, 3 or 4 weeks after T1. No significant changes were found in the levels of F0N0, bisection or galactosylation over time, but the amount of sialylation was significantly reduced between T1 and week-4 samples (Extended Data Fig. 4a). This suggested that infection triggered an acute increase in Fc sialylation that declined over time as new IgGs were produced, but this was not the case for other glycoforms. In contrast to SARS-CoV-2 infection, it was previously observed that acute dengue viral infections can trigger production of highly afucosylated IgGs that decline in the weeks following infection12. To further probe whether anti-RBD Fc glycoforms were associated with SARS-CoV-2 viral load, we performed correlation analyses between anti-RBD Fc glycoforms and SARS-CoV-2 RNA levels from nasopharyngeal swabs taken during acute infection. We observed no correlation between any Fc glycoforms and viral RNA load as determined by the cycle threshold value (Extended Data Fig. 4b). In all, these data did not support the hypothesis that SARS-CoV-2 virus infection regulated F0N0 Fc glycans during infection.
Fc afucosylation is elevated in male patients
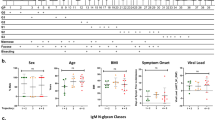
Studies have previously shown that antibody glycans may be influenced by age and sex13,14,15,16. We therefore performed a multivariate regression analysis on samples from patients who were hospitalized with COVID-19 from the Stanford Hospital Center (n = 30, 16 males and 14 females) to determine whether age and/or sex may have played a role in regulating the abundance of Fc glycans. We observed that sex was significantly correlated with Fc afucosylation (P = 0.0007) and age was not a confounding variable. Male patients had significantly higher levels of anti-RBD F0N0 Fc glycoforms than female patients (Fig. 4a). To determine the generalizability of this finding, we studied samples from a second cohort of patients who were hospitalized with COVID-19 in Northern California Kaiser Permanente hospitals (n = 81, 55 males and 26 females). In this larger cohort, male patients also had significantly elevated anti-RBD F0N0 Fc glycoforms compared to females (P = 0.019; Fig. 4b). A similar sex-associated difference in levels of afucosylation was not observed in patients with mild COVID-19 (n = 27, 13 males and 14 females; Fig. 4c). No other Fc glycoforms were segregated by sex in any of the cohorts (Extended Data Fig. 4c–e). Further, age did not correlate with any of the characterized anti-RBD Fc glycoforms (Extended Data Fig. 5). Overall, these data show that male patients with severe COVID-19 produced higher levels of afucosylated, anti-RBD IgG1 compared to females.
a,b, The level of the afucosylated anti-RBD IgG1 was significantly higher in hospitalized males (M) compared to hospitalized females (F) in two different cohorts from (a) Stanford Hospital Center (n = 30, M = 16 and F = 14; P = 0.0007) and (b) Kaiser Permanente hospitals of Northern California (n = 81, M = 55 and F = 26; P = 0.0190). c, Significant sex-associated differences in afucosylation levels of IgG1 were not present in patients with mild COVID-19 (outpatients; n = 27, M = 13 and F = 14). Violin plots in a–c show the distribution of sample values along with median (solid lines) and upper and lower quartile (dashed lines) values. P values were calculated using a two-tailed unpaired t-test with Welch’s correction. *P ≤ 0.05 and ***P ≤ 0.001.
Afucosylated SARS-CoV-2 immune complexes can promote FcγRIIIa interactions and inflammatory cytokine production
Afucosylation of IgG1 Fc confers five- to ten-fold higher affinity for the activating FcγRIIIa on a monomeric basis (and higher affinity in the context of a multivalent complex), compared to fucosylated IgG1 ref. 17). FcγRIIIa is present on monocytes, macrophages and NK cells in healthy adults and can enhance cell activation, proinflammatory cytokine production and cytotoxic effector cell activity18,19. Thus, distinct levels of fucosylation within SARS-CoV-2 immune complexes would be expected to modulate their binding to FcγRIIIa and activation of ITAM signaling17. To determine whether differential IgG1 fucosylation levels impacted their capacity to bind FcγRIIIa, we measured the apparent dissociation constant of purified serum IgGs from patients with severe COVID-19 with a range of fucose levels to recombinant FcγRIIIa (F158). IgGs from patients with RBD IgG1 afucosylation levels of >20% conferred ~threefold higher affinity to FcγRIIIa compared to IgGs with medium afucosylation and five- to six-fold higher affinity compared to IgGs with low IgG1 afucosylation (<10%). Further, RBD IgG1 afucosylation levels correlated with the apparent dissociation constant of IgGs for FcγRIIIa (P < 0.0001; Fig. 5a,b). To further characterize this using homogeneous Fc structures, we expressed the anti-SARS-CoV-2 RBD monoclonal antibody CR3022 (refs. 20,21) as a fully fucosylated IgG1 or as a variant with 32.6% afucosylated Fc, as determined by mass spectrometry; these variants differed in the apparent dissociation constant for FcγRIIIa by ~sevenfold (Fig. 5b). Next, we performed an ELISA to measure binding by IgGs from individual sera to FcγRIIIa. A total of 38 sera were selected, representing the range of afucosylation in comparison to the sample set. Consistent with the binding results using purified IgGs, we observed that the amount of anti-RBD IgG1 afucosylation correlated with binding to FcγRIIIa (P < 0.0001; Fig. 5c).
a, Binding of polyclonal IgGs was determined by biolayer interferometry. Graphs show the overlay of binding traces for a donor from each group representing varying degrees of anti-RBD core afucosylation: low (0–10%), medium (10–20%) and high (>20%). The kinetic constants were obtained by evaluating the binding at multiple concentrations (3.3 μM followed by twofold dilutions) of the analyte as shown (solid circles). The fits are indicated by solid lines. The assay was performed twice, and representative traces from one experiment are shown. b, Line graph showing a strong positive correlation (Pearson’s correlation coefficient, r = 0.86) between the apparent dissociation constant (KD,app) of FcγRIIIa (CD16a) and anti-RBD IgG1 core afucosylation in patients with COVID-19 (n = 13) as determined by biolayer interferometry. The binding of monoclonal anti-RBD CR3022 with different core afucosylation levels is also shown (red). Data represent the mean ± s.e.m. c, Line graph showing the correlation between the level of anti-RBD IgG1 Fc afucosylation and binding to FcγRIIIa. Binding of serum antibodies (n = 38) to FcγRIIIa positively correlated with the degree of afucosylation (Pearson’s correlation coefficient, r = 0.64). Samples are representative of the range of Fc fucosylation compared to the sample set. d, Line graph showing the correlation between the level of anti-RBD IgG1 Fc afucosylation and immune-complex-mediated NK cell degranulation. The amount of degranulation as measured by the fold increase of CD107a+ NK cells versus control correlated positively with the degree of afucosylation of the anti-RBD IgG (Spearman’s correlation coefficient, r = 0.79). The assay was performed in duplicate with peripheral blood mononuclear cells (PBMCs) from three healthy donors, and mean data are shown. e,f, Highly afucosylated immune complexes elicited increased production of inflammatory cytokines IL-6, TNF and IL-1β. Immune complexes were formed using pooled polyclonal IgGs from patients with COVID-19 (IL-6: P = 0.0081 for 100 µg ml−1, P < 0.0001 for 20 µg ml−1 and P = 0.0014 for 4 µg ml−1; TNF: P = 0.0042 for 100 µg ml−1, P = 0.0004 for 20 µg ml−1 and P = 0.0042 for 4 µg ml−1; IL-1β: P = 0.0068 for 100 µg ml−1, P = 0.0065 for 20 µg ml−1 and P = 0.0198 for 4 µg ml−1) (e) or recombinant IgG1 monoclonal antibody CR3022 with distinct levels of afucosylation (IL-6: P = 0.0009 for 10 µg ml−1, P = 0.0001 for 2 µg ml−1 and P = 0.0017 for 0.4 µg ml−1; TNF: P = 0.0022 for 10 µg ml−1, P = 0.0008 for 2 µg ml−1 and P = 0.0022 for 0.4 µg ml−1; IL-1β: P = 0.0152 for 10 µg ml−1 and P = 0.0231 for 2 µg ml−1). The assays were performed in duplicate with monocytes from three healthy donors and data represent the mean ± s.e.m. P values were calculated by two-tailed paired t-tests comparing high and low afucosylated immune complexes at each antibody concentration. *P ≤ 0.05, **P ≤ 0.01, ***P ≤ 0.001 and ****P ≤ 0.0001.
To determine whether these binding differences were physiologically relevant to activation of primary immune cells that express FcγRIIIa, we next performed in vitro stimulation assays. We first assessed activation of NK cells by immune complexes composed of patient IgGs and RBD. NK cell degranulation, measured by CD107a+ staining, correlated with the abundance of anti-RBD afucosylation in immune complexes (P = 0.0279; Fig. 5d). We next measured the ability of immune complexes with different levels of Fc afucosylation to stimulate cytokine production by primary monocytes. Immune complexes were formed from pooled IgGs mixed with SARS-CoV-2 pseudoparticles; these complexes were added to primary cells from separate donors. IgG pools were derived from patients with high (>20%) or low (<10%) anti-RBD IgG1 afucosylation and were matched for SARS-CoV-2 IgG binding titer (Extended Data Fig. 6). Highly afucosylated immune complexes triggered increased production of proinflammatory cytokines, principally IL-6, TNF and IL-1β, compared to immune complexes with low afucosylation (Fig. 5e and Extended Data Fig. 7a). To better define the role of Fc fucosylation alone on the differential activation of monocytes, we performed this assay with immune complexes made from SARS-CoV-2 pseudoparticles along with monoclonal anti-RBD CR3022 variants that differed only in Fc fucosylation, as described above. This modification had no impact on anti-spike binding (Extended Data Fig. 6). Cytokines including IL-6, TNF and IL-1β were significantly enhanced after incubation of monocytes with afucosylated monoclonal antibody CR3022 complexes (Fig. 5f and Extended Data Fig. 7b).
Overall, differences in core IgG1 afucosylation impacted FcγRIIIa binding and, within SARS-CoV-2 immune complexes, enrichment of afucosylated Fc structures could promote the production of cytokines including IL-6, TNF and IL-1β by primary monocytes. Thus, patients with severe COVID-19 were more likely to produce a proinflammatory form of IgG antibody (Extended Data Fig. 6).
Discussion
These studies show that specific proinflammatory antibody forms, characterized by IgG3 and IgG1 with F0N0 glycoform modification are elevated in more patients with severe COVID-19, in contrast to those with mild symptoms and seropositive children. We also found that male sex, in particular, was associated with high Fc afucosylation in severe COVID-19. Because previous studies in healthy adults have not found a sex-associated variation in IgG1 afucosylation, this correlation may arise from additional variables that were not identified in this study13,14,15.
Low-affinity activating FcγR pathways (FcγRIIa and FcγRIIIa) are often required for potent immunity mediated by passively transferred antibodies against viral pathogens22 and, indeed, one or both of these receptors likely has a role in protective antibody functions against SARS-CoV-2. The balance of activating to inhibitory FcγR signaling that promotes protective immunity and the thresholds for activating FcγR signals that may promote inflammatory pathology will be an important topic for future investigations into immunity against SARS-CoV-2. Overall, the data shown here raise the possibility that immune-complex-mediated activation of inflammatory FcγR pathways and the associated cytokine production is one ‘hit’ that can promote progression to severe COVID-19. A limitation of the present study is the absence of in vivo experimental data to define this because of the current lack of a humanized FcγR animal model for COVID-19 that would enable such mechanistic studies. In vivo models with well-characterized FcγR–IgG interactions and signaling outcomes will be essential for defining the role of antibodies in SARS-CoV-2 immunity and disease. Interestingly, previous studies have found high levels of afucosylated IgG1 in patients with severe dengue viral infections, and elevated maternal anti-dengue afucosylation predicted the risk for dengue disease in their infants10,12. Mechanistic studies have shown that afucosylated anti-dengue immune complexes and the associated enhancement in FcγRIIIa ITAM signaling can modulate diverse aspects of dengue virus–host interactions10,12.
These studies show that production of proinflammatory IgG antibodies was more common in severe cases of COVID-19. Future longitudinal studies including analysis of samples before and after infection will be needed to determine whether these Fc structures can be used as a preinfection biomarker for risk of progression to severe COVID-19.
Methods
Cloning, expression and protein purification
The His6-tagged SARS-CoV-2 RBD construct was a kind gift from F. Krammer (Icahn School of Medicine at Mount Sinai). The full-length recombinant SARS-CoV-2 spike protein (residues 1–1,208; GenBank no. MN908947, protein no. QHD43416.1) construct was designed with the following modifications: two well-characterized proline substitutions (K986P and V987P)23; a four amino acid substitution to remove the furin cleavage site (RRAR → GSAS) to stabilize the prefusion conformation24; a synthetic trimerization motif—the globular, β-rich ‘foldon’ from T4 fibritin to promote oligomerization in lieu of the native transmembrane domain; a human rhino virus 3C protease cleavage site; a C-terminal His8 tag. Mammalian codon-optimized gene fragments were synthesized (Integrated DNA Technologies) and cloned using Gibson Assembly (New England BioLabs) into a CMV/R promoter-driven mammalian expression vector between XbaI and BamHI restriction sites.
Both the constructs were transiently transfected into Expi293F cells (Thermo Fisher Scientific) per the manufacturer’s recommendations. Briefly, Expi293F cells at a density of 3 × 106 viable cells per ml maintained in Expi293 expression medium (Thermo Fisher Scientific) were transfected with expression plasmids complexed with ExpiFectamine 293 transfection reagent. Then, 18 h after transfection, the cells were supplemented with a cocktail of transfection enhancers. The cultures were incubated for 4 d, following which the culture supernatants were collected by centrifugation for protein purification. The supernatants were incubated with PBS (Gibco)-equilibrated Ni-nitrilotriacetic acid resin (GE Healthcare) for 2 h at 4 °C under mild mixing conditions to facilitate binding. The proteins were subsequently eluted using 500 mM imidazole (in PBS, pH 7.4) under gravity flow. The eluted fractions were pooled, buffer exchanged into PBS (pH 7.4) and concentrated using Amicon Ultra centrifugal units (EMD Millipore) to a final concentration of ~1 mg ml−1. Protein purity was assessed by SDS–PAGE. Size exclusion chromatography was used to determine the oligomeric state of the purified proteins under non-denaturing conditions at room temperature (RT) on a Superdex-200 analytical gel filtration column, and data were acquired on UNICORN 7 software (Cytiva). For molecular weight estimations, the column was calibrated using broad range molecular weight markers (Cytiva; Supplementary Fig. 1).
Monoclonal antibody CR3022 (ref. 21) was a kind gift from I. Wilson (The Scripps Research Institute) and was produced in Expi293F cells, as described above. For production of afucosylated monoclonal antibody CR3022, 200 μM of fucosyl transferase inhibitor 2F-peracetyl-fucose (Sigma Aldrich) was added to the culture after transfection. Supernatants were collected 5 d after transfection, and antibody purifications were performed over Protein G Sepharose 4 Fast Flow resin (GE Healthcare). Antibodies were buffer exchanged into PBS at pH 7.4 and concentrated using Amicon Ultra centrifugal units (EMD Millipore).
ELISAs
Screening ELISA
A rapid, high-throughput screening ELISA was performed on a total of 789 pediatric samples to test seropositivity following a modified version of a protocol described previously. Briefly, round-bottom 96-well plates (Immunolon 2HB; Thermo Scientific) were coated with 50 μl of RBD at 2 μg ml−1 in PBS for 1 h at RT. Next, the plates were blocked for 1 h with 3% non-fat milk in PBS with 0.1% Tween 20 (PBST). All serum samples from patients with COVID-19, the pediatric cohort and the negative controls were heated at 56 °C for 1 h, aliquoted and stored at −80 °C. For the first round of screening, all samples were diluted at 1:50 in 1% non-fat milk in PBST. Next, 50 μl of the diluted sera was added to each well and incubated for 2 h at RT. Following primary incubation with the sera, 50 μl of horseradish peroxidase (HRP)-conjugated goat anti-human IgG Fab (SouthernBiotech) at a dilution of 1:5,000 was added and incubated for 1 h at RT. The plates were developed by adding 50 μl per well of the chromogenic substrate 3,3′,5,5′-tetramethylbenzidine (TMB) solution (MilliporeSigma). The reaction was stopped with 0.2 N sulfuric acid (Sigma), and absorbance was measured at 450 nm (SPECTRAmax 250; Molecular Devices). The plates were washed five times with PBST between each step, and an additional wash with PBS was completed before developing the plates. Samples were considered seropositive against RBD if their absorbance value was greater than the mean plus four standard deviations of all negative controls (n = 130).
Validation ELISA of seropositive pediatric samples
The serum samples from the pediatric cohort, which showed seropositivity against RBD were validated by a second round of screening against the full-length SARS-Cov-2 spike protein. As described above, plates were coated with 50 μl of 2 μg ml−1 spike protein in PBS. Following blocking and washing, the plates were incubated with a series of fivefold dilutions of RBD-positive sera (50 μl) starting at 1:50 for 2 h at RT. All the subsequent steps were followed as described above. Sera from children and patients with COVID-19 were considered positive if they reached a threshold of the average value of 130 historical negative controls plus six standard deviations. The specificity of the assay was also tested on control sera from 12 participants with documented seasonal coronavirus infections collected in early 2019.
Isotyping by ELISA
Sera were diluted fivefold starting at 1:50, and ELISAs were performed as described above. The various secondary antibodies used for isotyping were 1:5,000 dilutions of HRP-conjugated goat anti-human IgG Fc (SouthernBiotech), mouse anti-human IgM (SouthernBiotech) and goat anti-human IgA Fc (SouthernBiotech).
CD16a ELISA
Human recombinant CD16a (Sino Biological) was immobilized at 3 μg ml−1 (50 μl per well) in PBS at 4 °C overnight, followed by 1 h of blocking with 3% non-fat milk in PBST. Next, 50 μl of 1:50 diluted sera from seropositive children (n = 11), PCR+ and seropositive patients (n = 22) and healthy controls (n = 5) was added to each well and incubated for 2 h at 37 °C. Subsequently, the plates were incubated for 1 h at 37 °C with 1:5,000 dilution of HRP-conjugated goat anti-human IgG Fc (SouthernBiotech) secondary antibody, developed as described previously with TMB, and absorbance was recorded at 450 nm.
Clinical cohorts and samples
Remnant sera from pediatric participants and from patients with PCR+ COVID-19 were obtained from Kaiser Permanente hospitals in Northern California. The sera were collected for a variety of clinical tests at one of 75 distinct hospitals or outpatient clinics across 17 counties in Northern California between 30 March 2020 to 19 April 2020. Serum samples from patients with PCR+ COVID-19 were also obtained from the Stanford ICU Biobank (protocol no. 28205) or from Stanford University (protocol no. NCT04331899)25. Characterization of these samples was performed under a protocol approved by the Institutional Review Board of Stanford University (protocol no. 55718).
Samples from people with seasonal coronavirus infections were collected at the University of Chicago. Samples were de-identified sera of healthcare workers that had respiratory illnesses, were swabbed and tested positive for common-cold coronaviruses in 2019 (University of Chicago protocol no. 09-043-A).
Historical controls and healthy controls
Thirty participants from a US cohort were enrolled at the Rockefeller University Hospital in New York City in 2012 in accordance with a protocol approved by the Institutional Review Board of Rockefeller University (protocol no. TWA-0804), in compliance with the International Conference on Harmonization Good Clinical Practice guidelines, and the protocol was registered on https://www.clinicaltrials.gov (NCT01967238). Blood samples were drawn from healthy adult volunteers between the ages of 18–64 years. Fifty samples were obtained from a Ugandan cohort of women and children enrolled in PROMOTE (NCT02163447), a randomized clinical trial of novel antimalarial chemoprevention regimens in Eastern Uganda26. The study was approved by the Institutional Review Boards of the Makerere University School of Biomedical Sciences, the Uganda National Council for Science and Technology and the University of California San Francisco. Written informed consent was obtained from all study participants. Fifty samples were obtained from children under 18 years of age enrolled in a study of acute febrile illness in Nepal. The study was approved by the Nepal Health Research Council, Kathmandu University Institutional Review Board and Stanford University Institutional Review Board (protocol no. 29992).
IgG Fc glycan and IgG subclass analysis
Methods for relative quantification of Fc glycans and IgG subclasses have been previously described11,12. Briefly, IgGs were isolated from serum by protein G purification. Antigen-specific IgGs were isolated on NHS agarose resin (Thermo Fisher; 26196) coupled to the protein of interest. Following tryptic digestion of purified IgG bound to antigen-coated beads, nanoscale liquid chromatography (nanoLC) coupled to tandem mass spectrometry analysis for characterization of glycosylation sites was performed on an UltiMate 3000 nanoLC (Dionex) coupled with a hybrid triple-quadrupole linear ion trap mass spectrometer, the 4000 Q Trap (SCIEX). Mass spectrometric data acquisition was performed using Analyst 1.6.1 software (SCIEX) for precursor ion scan triggered information-dependent acquisition (IDA) analysis for initial discovery-based identification.
For quantitative analysis of the glycoforms at the N297 site of IgG1, multiple-reaction monitoring (MRM) analysis for selected target glycopeptides was applied using the nanoLC-4000 Q Trap platform to the samples which were digested with trypsin. The m/z value of four charged ions for all different glycoforms as Q1 and the fragment ion at a m/z value of 366.1 as Q3 for each of the transition pairs were used for MRM assays. A native IgGs tryptic peptide (131-GTLVTVSSASTK-142) with transition pair of 575.9+2/780.4 was used as a reference peptide for normalization. IgG subclass distribution was quantitatively determined by nanoLC–MRM analysis of tryptic peptides following removal of glycans from purified IgGs with PNGase F. Here, the m/z value of fragment ions for monitoring transition pairs was always larger than that of their precursor ions to enhance the selectivity for unmodified targeted peptides and the reference peptide. All raw MRM data were processed using MultiQuant 2.1.1 (SCIEX). All MRM peak areas were automatically integrated and inspected manually. In instances where the automatic peak integration using MultiQuant failed, manual integration was performed using the MultiQuant software.
Binding affinity measurements using biolayer interferometry
Total IgGs were purified from sera of 13 patients with COVID-19 Protein G beads. The binding affinities of patient IgGs and high and low afucosylated CR3022 monoclonal antibodies were determined by biolayer interferometry using an OctetQK instrument (Pall ForteBio). Human recombinant CD16a (Sino Biological) was captured onto the amine reactive second-generation (AR2G) biosensors using the amine reactive second-generation reagent kit (Pall ForteBio). The CD16a bound sensors were dipped into a concentration series (3.33 μM, 1.7 μM, 0.832 μM and 0.33 μM) of IgGs in PBST to determine the binding kinetics. An equal number of unliganded biosensors dipped into the analytes served as controls for referencing. The traces were processed using ForteBio Data Analysis Software version 8.0.3.5 and corrected for nonspecific binding. The data were fitted globally to a simple 1:1 Langmuir interaction model.
NK cell degranulation assay
PBMCs were isolated from whole blood collected from healthy blood donors after plateletpheresis (Stanford Blood Center) using SepMate isolation tubes (STEMCELL). Cells were plated in a 96-well round-bottom plate (CELLSTAR) at a density of 3 × 106 cells per ml of complete RPMI1640 medium supplemented with 1× penicillin–streptomycin–glutamine, 1 mM sodium pyruvate and 1× MEM non-essential amino acids, 10% heat-inactivated fetal bovine serum (Gibco) and 1 ng ml−1 IL-15 (STEMCELL) and rested overnight at 37 °C in a 5% CO2 incubator (Panasonic). The following morning, cell culture medium was replaced with complete RPMI containing anti-CD107a (BioLegend; clone H4A3). PBMCs were promptly stimulated for 6 h at 37 °C with immune complexes formed by incubating purified patient IgG with SARS-CoV-2 RBD protein at a molar ratio of 30:1 for 1 h at RT. Approximately 1 h into stimulation, culture medium was supplemented with 1× brefeldin A (BioLegend) for the remaining 5 h of culture. Cells were then isolated, stained for cell viability using a Live/Dead Fixable Staining Kit (Thermo Fisher; 1:60 dilution), as well as CD3 (clone OKT3), CD11c (clone S-HCL-3), CD14 (clone 63D3), CD16 (clone 3G8), CD56 (clone HCD56) and HLA-DR (clone L243) surface markers (BioLegend) (all 1:30 dilution). After staining, cells were fixed and acquired using an Attune NxT flow cytometer (Invitrogen). NK cells were defined as viable CD3−CD14−CD16+CD56+HLA-DR− cells (Supplementary Fig. 1). NK cell degranulation was measured and reported as the fold change of NK cells positive for CD107a compared to control.
Monocyte stimulation and cytokine measurements
Monocytes were isolated from healthy donor blood (Stanford Blood Center) using RosetteSep Human Monocyte Enrichment Kit (STEMCELL) per the manufacturer’s instructions. Monocytes were cultured at a density of 2 × 106 cells per ml in RPMI 1640 medium supplemented with 1× non-essential amino acids, sodium pyruvate, penicillin–streptomycin–glutamine (Gibco) and 10% fetal bovine serum (GE Healthcare Life Sciences). Immune complexes were formed by incubating a dilution series of IgGs from patients with COVID-19 or anti-spike CR3022 monoclonal antibodies to SARS-CoV-2 spike-expressing a ΔG-VSV pseudovirus for 1 h at RT. Monocytes were incubated with the various immune complexes or the pseudovirus for only 18 h at 37 °C in a 5% CO2 incubator. After 18 h, cell-free supernatants were collected and proinflammatory cytokine concentrations were measured using a LEGENDplex bead array (BioLegend) according to the manufacturer’s instructions.
Statistical analysis
R statistical package version 1.2.1335 was used to generate the radar plots and perform multivariate linear regression analysis. All other data were analyzed with GraphPad Prism 8.0 software. Investigators were blinded to study participants’ diagnoses during screening; patients with COVID-19 and children were not known by investigators at the time of ELISA screening for RBD reactivity of serum or by investigators involved in relative quantitation of Fc glycoforms and IgG subclasses by mass spectrometry.
Reporting Summary
Further information on research design is available in the Nature Research Reporting Summary linked to this article.
Data availability
All raw data are available from the corresponding author on reasonable request. The reference SARS-CoV-2 spike protein sequence was obtained from the NCBI database (https://www.ncbi.nlm.nih.gov/protein/1791269090).
References
Bournazos, S., Wang, T. T., Dahan, R., Maamary, J. & Ravetch, J. V. Signaling by antibodies: recent progress. Annu. Rev. Immunol. 35, 285–311 (2017).
Wang, T. T. IgG Fc glycosylation in human immunity. Curr. Top. Microbiol Immunol. 423, 63–75 (2019).
Nimmerjahn, F. & Ravetch, J. V. Fc receptors as regulators of immunity. Adv. Immunol. 96, 179–204 (2007).
Nimmerjahn, F. & Ravetch, J. V. Divergent immunoglobulin G subclass activity through selective Fc receptor binding. Science 310, 1510–1512 (2005).
Liu, W. et al. Detection of COVID-19 in children in early January 2020 in Wuhan, China. N. Engl. J. Med. 382, 1370–1371 (2020).
Xu, Y. et al. Characteristics of pediatric SARS-CoV-2 infection and potential evidence for persistent fecal viral shedding. Nat. Med. 26, 502–505 (2020).
Bi, Q. et al. Epidemiology and transmission of COVID-19 in 391 cases and 1,286 of their close contacts in Shenzhen, China: a retrospective cohort study. Lancet Infect. Dis. 20, 911–919 (2020).
Dong, Y. et al. Epidemiology of COVID-19 among children in China. Pediatrics 145, e20200702 (2020).
Stadlbauer, D. et al. SARS-CoV-2 seroconversion in humans: a detailed protocol for a serological assay, antigen production and test setup. Curr. Protoc. Microbiol. 57, e100 (2020).
Thulin, N. K. & Wang, T. T. Maternal anti-dengue IgG fucosylation predicts susceptibility to dengue disease in infants. Cell Rep. 31, 107642 (2020).
Wang, T. T. et al. Anti-HA glycoforms drive B cell affinity selection and determine influenza vaccine efficacy. Cell 162, 160–169 (2015).
Wang, T. T. et al. IgG antibodies to dengue enhanced for FcγRIIIA binding determine disease severity. Science 355, 395–398 (2017).
Bakovic, M. P. et al. High-throughput IgG Fc N-glycosylation profiling by mass spectrometry of glycopeptides. J. Proteome Res. 12, 821–831 (2013).
Chen, G. et al. Human IgG Fc-glycosylation profiling reveals associations with age, sex, female sex hormones and thyroid cancer. J. Proteomics 75, 2824–2834 (2012).
de Haan, N., Reiding, K. R., Driessen, G., van der Burg, M. & Wuhrer, M. Changes in healthy human IgG Fc-glycosylation after birth and during early childhood. J. Proteome Res. 15, 1853–1861 (2016).
Kapur, R. et al. A prominent lack of IgG1-Fc fucosylation of platelet alloantibodies in pregnancy. Blood 123, 471–480 (2014).
Li, T. et al. Modulating IgG effector function by Fc glycan engineering. Proc. Natl Acad. Sci. USA 114, 3485–3490 (2017).
Kramer, P. R., Winger, V. & Reuben, J. PI3K limits TNF-α production in CD16-activated monocytes. Eur. J. Immunol. 39, 561–570 (2009).
Bournazos, S., Wang, T. T. & Ravetch, J. V. In Myeloid Cells in Health and Disease: A Synthesis (Ed. Gordon, S.) Ch. 22 (ASM Press, 2016).
Yuan, M. et al. A highly conserved cryptic epitope in the receptor-binding domains of SARS-CoV-2 and SARS-CoV. Science 368, 630–633 (2020).
ter Meulen, J. et al. Human monoclonal antibody combination against SARS coronavirus: synergy and coverage of escape mutants. PLoS Med. 3, e237 (2006).
Bournazos, S. & Ravetch, J. V. Fcγ receptor pathways during active and passive immunization. Immunol. Rev. 268, 88–103 (2015).
Pallesen, J. et al. Immunogenicity and structures of a rationally designed prefusion MERS-CoV spike antigen. Proc. Natl Acad. Sci. USA 114, E7348–E7357 (2017).
Wrapp, D. et al. Cryo-EM structure of the 2019-nCoV spike in the prefusion conformation. Science 367, 1260–1263 (2020).
Wilk A. J. et al. A single-cell atlas of the peripheral immune response in patients with severe COVID-19. Nat. Med. 26, 1070–1076 (2020).
Jagannathan, P. et al. Dihydroartemisinin-piperaquine for intermittent preventive treatment of malaria during pregnancy and risk of malaria in early childhood: a randomized controlled trial. PLoS Med. 15, e1002606 (2018).
Acknowledgements
Support was received from Stanford University, the Chan Zuckerberg Biohub and the Searle Scholars Program. Research reported in this publication was supported by Fast Grants and the CEND COVID Catalyst Fund. Additional funding was received from the National Institute of Allergy and Infectious Diseases of the National Institutes of Health under award numbers U19AI111825, U54CA260517, R01AI139119, R01AI130398 and R01AI127877. Clinical samples were obtained with support from the Rockefeller University Center for Clinical and Translational Science (grant no. UL1 TR001866). The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health. T.T.W. thanks C. Kopel for exceptional technical support.
Author information
Authors and Affiliations
Contributions
T.T.W., S.C., K.E. and J.G. conceived and/or designed the experiments. T.T.W., S.C., K.E., R.S., S.Z., V.M., A.S.B., M.M.X., J.G., C.B., N.K., S.P., J.R.A., J.M.S., T.M., C.A.B., P.C.W., T.D.P., S.D.B., U.S., B.A.P., K.C.N., H.D., M.J.M., J.K.T., G.S.T. and P.J. were involved in data acquisition and/or analysis and/or interpretation. S.C. and T.T.W. wrote the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Peer review information Nature Immunology thanks Robert Anthony and the other, anonymous, reviewer(s) for their contribution to the peer review of this work. Zoltan Fehervari was the primary editor on this article and managed its editorial process and peer review in collaboration with the rest of the editorial team.
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data
Extended Data Fig. 1 ROC curves with other characterized IgG1 glycans.
Receiver operating characteristic (ROC) curve for anti-RBD IgG1 IgG1 bisection (blue), galactosylation (green) and sialylation (yellow) from all hospitalized (n = 43) and mild (n = 18) COVID-19 patients showed these glycans were not predictors of COVID-19 severity. Area under the curve (AUC) for bisection 0.54[95% CI (0.38-0.69, p = 0.6357), galactosylation 0.61[0.42–0.81, p = 0.1872 and sialylation 0.60[95% CI (0.40–0.79, p = 0.2245).
Extended Data Fig. 2 Regulation of Fc glycoforms.
(a) Longitudinal analysis of the levels of anti-RBD IgG1 Fc afucosylation from COVID-19 patients at various time points. T1 represents the first draw with a second time point at week 2 (n = 5), week 3 (n = 5) or week 4 (n = 11). Anti-RBD IgG1 from COVID-19 patients were characterized for Fc afucosylation (F0N0), bisection (N), sialylation (S) and galactosylation (GS0). None of the glycan levels were significantly different between T1 and subsequent time-points post infection. P values were calculated using two-tailed Wilcoxon matched-pairs signed rank test. (b) There was no correlation (Pearson’s correlation coefficient (r) was calculated, and two-tailed p and r values have been reported) between the viral load and the abundance of any of the glycans on anti-RBD IgG1 (n = 25). (c, d) The levels of the various glycans were not significantly different amongst male and female hospitalized COVID-19 patients in two cohorts (Blue-Kaiser Permanente (n = 81, M = 55, F = 26), Orange-Stanford hospital (n = 30, F = 14, M = 16)) or in (e) mild (green) COVID-19 patients (n = 27, F = 14, M = 13). Violin plots in (c), (d) and (e) show the distribution of sample values along with median (solid lines) and quartile (broken lines) values. P values were calculated using a two-tailed unpaired t test with Welch’s correction.
Extended Data Fig. 3 Correlation between age and abundance of IgG1 glycoforms.
Age of COVID-19 patients (n = 107) did not correlate with abundance of any of the Fc glycoforms. Pearson’s correlation coefficient (r) and two-tailed p values have been reported.
Extended Data Fig. 4 Binding by differentially afucosylated IgGs to SARS-CoV-2 S protein.
The level of IgG afucosylation did not impact binding to spike protein both for (a) pooled IgG from patients or (b) mAb 3022. The assays were performed in duplicate and mean data and standard deviation (SD) have been graphed.
Extended Data Fig. 5 Cytokine production by immune complex-stimulated monocytes.
Monocytes stimulated with highly afucosylated immune complexes made from both (a) patient IgG pools (IL8: p = 0.0196 for 100 µg/ml, p = 0.0002 for 20 µg/ml. IL10: p < 0.0001 for 100 µg/ml, p < 0.0001 for 20 µg/ml and p = 0.0019 for 4 µg/ml. GM-CSF: p = 0.0036 for 100 µg/ml, p < 0.0001 for 20 µg/ml and p = 0.003 for 4 µg/ml) and (b) mAb 3022 (IL8: p = 0.001 for 10 µg/ml, p = 0.0011 for 2 µg/ml. IL10: p = 0.0045 for 10 µg/ml, p = 0.0112 for 2 µg/ml and p = 0.0021 for 0.4 µg/ml. GM-CSF: p = 0.0007 for 10 µg/ml, p < 0.0001 for 2 µg/ml and p = 0.0036 for 0.4 µg/ml) produced higher amounts of multiple proinflammatory cytokines as compared to immune complexes with low afucosylation. The assays were performed in duplicate with monocytes from three healthy donors and mean data and standard error of the mean (SEM) has been graphed. P-values between high and low afucosylated immune complexes at each antibody concentration were calculated by two-tailed paired t-tests. *P ≤ 0.05, **P ≤ 0.01, ***P ≤ 0.001, ****P ≤ 0.0001.
Extended Data Fig. 6 Schematic representation of the role of IgG afucosylation in COVID-19.
(a) Severe COVID-19 patients were more likely to have elevated levels of afucosylated IgG1 as compared to patients with mild disease. (b) Immune complexes formed from these high afucosylated antibodies have stronger binding to low affinity activating FcγRIIIa on surface of innate immune cells and thus more ITAM signaling. This leads to increased production of pro-inflammatory cytokines.
Supplementary information
Supplementary Information
Supplementary Fig. 1
Rights and permissions
About this article
Cite this article
Chakraborty, S., Gonzalez, J., Edwards, K. et al. Proinflammatory IgG Fc structures in patients with severe COVID-19. Nat Immunol 22, 67–73 (2021). https://doi.org/10.1038/s41590-020-00828-7
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41590-020-00828-7
This article is cited by
-
Control of complement-induced inflammatory responses to SARS-CoV-2 infection by anti-SARS-CoV-2 antibodies
The EMBO Journal (2024)
-
Immunoglobulin G N-glycan markers of accelerated biological aging during chronic HIV infection
Nature Communications (2024)
-
Mechanism of glycoform specificity and in vivo protection by an anti-afucosylated IgG nanobody
Nature Communications (2023)
-
A highly potent human neutralizing antibody prevents vertical transmission of Rift Valley fever virus in a rat model
Nature Communications (2023)
-
Beyond neutralization: Fc-dependent antibody effector functions in SARS-CoV-2 infection
Nature Reviews Immunology (2023)