Abstract
The phylogenetic relationships and distribution of Ralstonia pseudosolanacearum and Ralstonia solanacearum sequevars were analyzed in 108 strains of Solanaceae in three mesoregions of the Pernambuco State, Brazil. These mesoregions are characterized by a semiarid climate and account for 88.20% of the area of the state. Ralstonia pseudosolanacearum (phylotype I, biovar 3) was predominant in Agreste, and R. solanacearum (phylotype II, biovar 1) was predominant in São Francisco and Sertão. Two haplotypes of R. pseudosolanacearum were identified by analysis of the haplotype network of the partial egl gene sequences and were phylogenetically related to sequevars I-17 and I-18. Five haplotypes of R. solanacearum were identified; two haplotypes were related to sequevar IIA-50, and three haplotypes were related to new sequevars IIA-61, IIA-62, and IIA-63. In Brazil, sequevars I-17, IIA-61, IIA-62, and IIA-63 were detected exclusively in the semiarid climate of Pernambuco, unlike sequevars I-18 and IIA-50, which were detected in other regions of Brazil. Furthermore, R. pseudosolanacearum (phylotype I) and sequevar IIA-50 were detected in the Sertão mesoregion. Sequevar I-18, IIA-61, and IIA-62 strains were prevalent in the Agreste mesoregion, and sequevars I-17 and IIA-50 were prevalent in the São Francisco mesoregion. The virulence variability of the strains toward tomato was related to species and sequevars. Introduced R. pseudosolanacearum strains of sequevar I-18 had higher estimated virulence compared to that of the native R. solanacearum strains of sequevar IIA-50. This is the first study that analyzes the variability of Ralstonia spp. in the semiarid region of Brazil.



Similar content being viewed by others
References
Ab’ Saber, A. N. (1967). Os Domínios Morfoclimáticos e Províncias Fitogeográficas do Brasil. São Paulo, SP: USP.
Albuquerque, G. M. R., Santos, L. A., Felix, K. C. S., Rollemberg, C. L., Silva, A. M. F., Souza, E. B., Cellier, G., Prior, P., & Mariano, R. L. R. (2014). Moko disease causing strains of Ralstonia solanacearum from Brazil extend known diversity in paraphyletic phylotype II. Phytopathology, 104(11), 1175–1182. https://doi.org/10.1094/PHYTO-12-13-0334-R.
Albuquerque, G. M. R., Silva, A. M. F., Silva, J. R., Ferraz, E., Silva, M. C. L., & Souza, E. B. (2015). Levantamento da murcha bacteriana em solanáceas no estado de Pernambuco. http://www.eventosufrpe.com.br/2015/cd/resumos/R0762-1.html. Accessed 10 June 2020.
Albuquerque, G. M. R., Silva, A. M. F., Silva, J. R., Gama, M. A. S., & Mariano, R. L. R. (2016). First report of bacterial wilt caused by Ralstonia pseudosolanacearum on Eruca vesicaria subsp sativa in Brazil. Plant Disease, 8(11), 2319. https://doi.org/10.1094/PDIS-04-16-0420-PDN.
Bergsma-Vlami, M., Van de Bilt, J. L. J., Tjou-Tam-Sin, N. N. A., Westenberg, M., Meekes, E. T. M., Teunissen, H. A. S., & Van Vaerenbergh, J. (2018). Phylogenetic Assingnment of Ralstonia pseudosolanacearum (Ralstonia solanacearum Phylotype I) isolated from Rosa spp. Plant Disease, 102(11), 2258–2267. https://doi.org/10.1094/PDIS-09-17-1345-RE.
Brasil-Min. Ministério da Integração Nacional. (2005). Nova Delimitação do Semi-árido brasileiro. http://www.cpatsa.embrapa.br/public_eletronica/downloads/OPB1839.pdf. Accessed 07 August 2019.
Carstensen, G. D., Venter, S. N., Wingfield, M. J., & Coutinho, T. A. (2017). Two Ralstonia species associated with bacterial wilt of Eucalyptus. Plant Patholology, 66(3), 393–403. https://doi.org/10.1111/ppa.12577.
Cellier, G., & Prior, P. (2010). Deciphering phenotypic diversity of Ralstonia solanacearum strains pathogenic to potato. Phytopathology, 100(11), 1250–1261. https://doi.org/10.1094/PHYTO-02-10-0059
Chesneau, T., Maignien, G., Boyer, C., Chéron, J.-J., Roux-Cuvelier, M., Vanhuffel, L., Poussier, S., & Prior, P. (2018). Sequevar diversity and virulence of Ralstonia solanacearum Phylotype I on Mayotte Island (Indian Ocean). Frontiers in Plant Science, 8, 2209. https://doi.org/10.3389/fpls.2017.02209/full.
Cho, H., Song, E.-S., Lee, Y. K., Lee, S., Lee, S.-W., Jo, A., Lee, B.-M., Kim, J.-G., & Hwang, I. (2018). Analysis of genetic and pathogenic diversity of Ralstonia solanacearum causing potato bacterial wilt in Korea. Plant Patholology Journal, 34(1), 23–34. https://doi.org/10.5423/PPJ.FT.09.2017.0203.
Costa, S. B., Ferreira, M. A. S. V., & Lopes, C. A. (2007). Diversidade patogênica e molecular de Ralstonia solanacearum da Região Amazônica brasileira. Fitopatologia Brasileira, 32(4), 285–294. https://doi.org/10.1590/S0100-41582007000400002.
Darriba, D., Taboada, G. L., Doallo, R., & Posada, D. (2012). JModelTest 2: More models, new heuristics and parallel computing. Nature Methods, 9(8), 772. https://doi.org/10.1038/nmeth.2109.
Elphinstone, J. G. (2005). The current bacterial wilt situation: A global overview. In C. Allen, P. Prior, & A. C. Hayward (Eds.), Bacterial wilt disease and the Ralstonia solanacearum species complex (pp. 9–28). St. Paul: APS Press.
Fegan, M., & Prior, P. (2005). In C. Allen, P. Prior, & A. C. Hayward (Eds.), Bacterial wilt disease and the Ralstonia solanacearum species complex (pp. 449-461). St. Paul: APS press.
Fegan, M., & Prior, P. (2006). Diverse members of the Ralstonia solanacearum species complex cause bacterial wilt of banana. Australasian Plant Pathology, 35(2), 93–101. https://doi.org/10.1071/AP05105.
Felix, K. C. S., Souza, E. B., Michereff, S. J., & Mariano, R. L. R. (2012). Survival of Ralstonia solanacearum in infected tissues of Capsicum annuum and in soils of the state of Pernambuco, Brazil. Phytoparasitica, 40(1), 53–62. https://doi.org/10.1007/s12600-011-0200-6.
Ferreira, D. F. (2011). Sisvar: um sistema computacional de análise estatística. Ciência e Agrotecnologia, 35(6), 1039–1042. https://doi.org/10.1590/S1413-70542011000600001.
Garcia, A. L., Lima, W. G., Souza, E. B., Michereff, S. J., & Mariano, R. L. R. (2013). Characterization of Ralstonia solanacearum causing bacterial wilt bell pepper in the state Pernambuco, Brasil. Journal of Plant Pathology, 95(2), 237–245. https://doi.org/10.4454/JPP.V95I2.032.
Guindon, S., & Gascuel, O. (2003). A simple, fast, and accurate algorithm to estimate large phylogenies by maximum likelihood. Systematic Biology, 52(5), 696–704. https://doi.org/10.1080/10635150390235520.
Guindon, S., Dufayard, J. F., Lefort, V., Anisimova, M., Hordijk, W., & Gascuel, O. (2010). New algorithms and methods to estimate maximum-likelihood phylogenies: assessing the performance of PhyML 3.0. Systematic Biology, 59(3), 307–321. https://doi.org/10.1093/sysbio/syq010.
Gutarra, L., Herrera, J., Fernandez, E., Kreuze, J., & Lindqvist-Kreuze, H. (2017). Diversity, pathogenicity, and current occurrence of bacterial wilt bacterium Ralstonia solanacearum in Peru. Frontiers in Plant Science, 8, 1221. https://doi.org/10.3389/fpls.2017.01221.
Hasegawa, M., Kishino, K., & Yano, T. (1985). Dating the human-ape splitting by a molecular clock of mitochondrial DNA. Journal of Molecular Evolution, 22(2), 160–174. https://doi.org/10.1007/BF02101694.
Hayward, A. C. (1994). Systematics and phylogeny of Pseudomonas solanacearum and related bacteria. In A. C. Hayward & G. L. Hartman (Eds.), Bacterial wilt: The disease and its causative agent, Pseudomonas solanacearum (pp. 123–135). Wallingford: CAB International.
Hfbrasil. (2019). Anuário 2018–2019 Hortifruti Brasil. https://www.hfbrasil.org.br/br/revista/acessar/completo/anuario-2018-2019.aspx. Accessed 08 August 2019.
Kelman, A. (1954). The relationship of pathogenicity in Pseudomonas solanacearum to colony appearance on a tetrazolium medium. Phytopathology, 44(12), 693–695.
Kumar, S., Stecher, G., & Tamura, K. (2016). MEGA7: Molecular evolutionary genetics analysis version 7.0 for bigger datasets. Molecular Biology and Evolution, 33(7), 1870–1874. https://doi.org/10.1093/molbev/msw054.
Li, Y., Feng, J., Liu, H., Wang, L., Hsiang, T., Li, X., & Huang, J. (2016). Genetic diversity and pathogenicity of Ralstonia solanacearum causing tobacco bacterial wilt in China. Plant Disease, 100(7), 1288–1296. https://doi.org/10.1094/PDIS-04-15-0384-RE.
Liu, Y., Wu, D., Liu, Q., Zhang, S., Tang, Y., Jiang, G., Li, S., & Ding, W. (2017). The sequevar distribution of Ralstonia solanacearum in tobacco-growing zones of China is structured by elevation. European Journal Plant Pathology, 147(3), 541–551. https://doi.org/10.1007/s10658-016-1023-6.
Lopes, C. A., & Duval, A. M. Q. (2007). Epidemiologia e controle das bacterioses das hortaliças. In L. Zambolim, C. A. Lopes, M. C. Picanço, & H. Costa (Eds.), Manejo Integrado de Doenças e Pragas de Hortaliças (pp. 115–162). Viçosa: Editora UFV.
Nielsen, L. W., & Haynes, F. L. (1960). Resistance in Solanum tuberosum to Pseudomonas solanacearum. American Potato Journal, 37(8), 260–267.
Opina, N., Tavner, F., Holloway, G., & Timmis, J. N. (1997). A novel method for development of species and strain-specific DNA probes and PCR primers for identifying Burkholderia solanacearum (formerly Pseudomonas solanacearum). Asia Pacific Journal of Molecular Biology and Biotechnology, 5(1), 19–33.
Pernambuco-Bde. Pernambuco Base de dados do estado. (2019). Divisão geopolítica: mesorregiões. http://www.bde.pe.gov.br/estruturacaogeral/mesorregioes.aspx. Accessed 07 August 2019.
Posada, D. (2003). Using JModelTest and PAUP* to select a model of nucleotide substitution. Current Protocols in Bioinformatics. https://doi.org/10.1002/0471250953.bi0605s00
Poussier, S., Trigalet-Demery, D., Vanderwalle, P., Goffinet, B., Luisetti, J., & Trigalet, A. (2000). Genetic diversity of Ralstonia solanacearum as assessed by PCR R-RFLP of the hrp gene region, AFLP and 16S rRNA sequence analysis, and identification of an African subdivision. Microbiology, 146(7), 1679–1692. https://doi.org/10.1099/00221287-146-7-1679.
Prior, P., Ailloud, F., Dalsing, B. L., Remenant, B., Sanchez, B., & Allen, C. (2016). Genomic and proteomic evidence supporting the division of the plant pathogen Ralstonia solanacearum into three species. BMC Genomics, 17, 90. https://doi.org/10.1186/s12864-016-2413-z.
Ravelomanantsoa, S., Vernière, C., Rieux, A., Costet, L., Chiroleu, F., Arribat, S., Cellier, G., Pruvost, O., Poussier, S., Robène, I., Guérin, F., & Prior, P. (2018). Molecular epidemiology of bacterial wilt in the Madagascar highlands caused by Andean (Phylotype IIB-1) and African (Phylotype III) Brown rot strains of the Ralstonia solanacearum species complex. Frontiers in Plant Science, 8, 2258. https://doi.org/10.3389/fpls.2017.02258.
Rodrigues, L. M. R., Destefano, S. A. L., Silva, M. J., Costa, G. G. L., & Maringoni, A. C. (2012). Characterization of Ralstonia solanacearum from Brazil using molecular methods and pathogenicity tests. Journal of Plant Pathology, 94(3), 505–516. https://doi.org/10.4454/JPP.FA.2012.052.
Ronquist, F., & Huelbecsenk, J. P. (2003). MrBayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics, 19(12), 1572–1574. https://doi.org/10.1093/bioinformatics/btg180.
Safni, I., Cleenwerck, I., De Vos, P., Fegan, M., Sly, L., & Kappler, U. (2014). Polyphasic taxonomic revision of the Ralstonia solanacearum species complex: proposal to emend the descriptions of Ralstonia solanacearum and Ralstonia syzygii and reclassify current R. syzygii strains as Ralstonia syzygii subsp. syzygii subsp. nov., R. solanacearum phylotype IV strains as Ralstonia syzygii subsp. indonesiensis subsp.nov., banana blood disease bacterium strains as Ralstonia syzygii subsp. celebesensis subsp. nov. and R. solanacearum phylotype I and III strains as Ralstonia pseudosolanacearum sp. nov. International Journal Systematic Evolutionary Microbiology, 64(9), 3087–3103. https://doi.org/10.1099/ijs.0.066712-0.
Saitou, N., & Nei, M. (1987). The neighbor-joining method: A new method for reconstructing phylogenetic trees. Molecular Biology and Evolution, 4(4), 406–425. https://doi.org/10.1093/oxfordjournals.molbev.a040454.
Santiago, T. R., Lopes, C. A., Caetano-Anolles, G., & Mizubuti, E. S. G. (2017). Phylotype and sequevar variability of Ralstonia solanacearum in Brazil, an ancient Centre of diversity of the pathogen. Plant Pathology, 66(3), 383–392. https://doi.org/10.1111/ppa.12586.
She, X., Yu, L., Lan, G., Tang, Y., & He, Z. (2017). Identification and genetic characterization of Ralstonia solanacearum species complex isolates from Cucurbita maxima in China. Frontiers in Plant Science, 8, 1794. https://doi.org/10.3389/fpls.2017.01794.
Silva, J. R. (2014). Diversidade de isolados de Ralstonia solanacearum das regiões norte e nordeste do Brasil. http://www.pgfitopat.ufrpe.br/sites/ww2.prppg.ufrpe.br/files/dissertacao_jessica_2014.pdf. Accessed 07 July 2019.
Staden, R., Beal, K. F., & Bonfield, J. K. (1998). The Staden package. In S. Misener & S. A. Krawetz (Eds.), Bioinformatics methods and protocols (pp. 115–130). New York: Humana.
Stulberg, M. J., & Huang, Q. (2016). A computer program for fast and easy typing of a partial endoglucanase gene sequence into genospecies and sequevars 1 & 2 of the Ralstonia solanacearum species complex. Journal of Microbiological Methods, 123, 101–107. https://doi.org/10.1016/j.mimet.2016.02.010.
Thano, P., & Akarapisan, A. (2018). Phylotype and sequevar of Ralstonia solanacearum which causes bacterial wilt in Curcuma alismatifolia Gagnep. Letters in Applied Microbiology, 66(5), 384–393. https://doi.org/10.1111/lam.12857.
Tompson, J. D., Higgine, D. G., & Gibson, T. J. (1994). Clustal W: Improving the sensivity of multiple sequence alignment through sequence weigting, position specifc gap penalties and weight matrix choice. Nucleic Acids Research, 22(22), 4673–4680. https://doi.org/10.1093/nar/22.22.4673.
Toukam, G. M. S., Cellier, G., Wicker, E., Guilbaud, C., Kahane, R., Allen, C., & Prior, P. (2009). Broad diversity of Ralstonia solanacearum strains in Cameroon. Plant Disease, 93(11), 1123–1130. https://doi.org/10.1094/PDIS-93-11-1123.
Wicker, E., Lefeuvre, P., de Cambiaire, J. C., Lemaire, C., Poussier, S., & Prior, P. (2012). Contrasting recombination patterns and demographic histories of the plant pathogen Ralstonia solanacearum inferred from MLSA. The ISME Journal, 6(5), 961–974. https://doi.org/10.1038/ismej.2011.160.
Xu, J., Pan, Z. C., Xu, J. S., Zhang, Z., Zhang, H., Zhang, L. Q., et al. (2009). Genetic diversity of Ralstonia solanacearum strains from China. European Journal of Plant Pathology, 125(4), 641–653. https://doi.org/10.1007/s10658-009-9512-5
Yahiaoui, N., Chéron, J.-J., Ravelomanantsoa, S., Hamza, A. A., Petrousse, B., Jeetah, R., Jaufeerally-Fakim, Y., Félicité, J., Fillâtre, J., Hostachy, B., Guérin, F., Cellier, G., Prior, P., & Poussier, S. (2017). Genetic diversity of the Ralstonia solanacearum species complex in the Southwest Indian Ocean Islands. Frontiers in Plant Science, 8, 2139. https://doi.org/10.3389/fpls.2017.02139.
Zhang, Y., & Qiu, S. (2016). Phylogenomic analysis of the genus Ralstonia based on 686 single-copy genes. Antonie Van Leeuwenhoek, 109(1), 71–82. https://doi.org/10.1007/s10482-015-0610-4.
Acknowledgments
The Fundação de Amparo à Ciência e Tecnologia do Estado de Pernambuco (FACEPE) for the granting of a researcher fixation grant to G.M.R. Albuquerque (Proc. BFP 0158-5.01/16). Extensionists, technicians and agronomists of the Agronomic Institute of Pernambuco (IPA) of Belém do São Francisco, Petrolina, and Salgueiro municipalitys, to Embrapa Semiarido and the Agricultural Defense Agency of Pernambuco (ADAGRO) for their collaboration in visits to the Solanaceae productions areas in São Francisco and Sertão mesoregions. To PhD. Gilles Cellier of Agence Nationale de Sécurité Sanitaire de l’Alimentation, de l’Environnement et du Travail from France by revision of new sequevars sequences.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Research involving human participants and/or animals
This study does not include any trial with human and/or animals.
Informed consent
Informed consent was obtained from participants included in the study.
Supplementary Information
Supplementary Fig. S1
Bacterial wilt in the semiarid climate of Pernambuco state, Northeast Brazil. A, Representation of the map of Brazil indicating the location of Pernambuco state (circle) and the region characterized by a semiarid climate (part colored in gray); B, delimitation of the municipalities and mesoregions (Agreste, São Francisco, and Sertão) in Pernambuco state with a semiarid climate where the Solanaceae with bacterial wilt was collected. (BMP 2730 kb)
Supplementary Fig. S2
Ralstonia spp. strains (n = 108) in the semiarid climate of Pernambuco state, Northeast Brazil. A, Ralstonia spp. strains distribution (%) by mesoregion; B, Ralstonia pseudosolanacearum and R. solanacearum strains distribution (%) by mesoregion. (BMP 2.54 kb)
ESM 3
(PNG 2.75 mb)
Supplementary Fig. S3
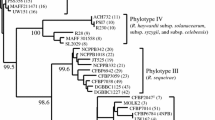
Phylogenetic reconstruction based on partial egl gene sequences from strains of Ralstonia pseudosolanacearum and R. solanacearum causing bacterial wilt in the semiarid climate of Pernambuco state, Brazil. This was conducted using the neighbor-joining method with the Jukes and Cantor algorithm and with 2000 bootstraps to test the tree topology in MEGA 7.0 software. Representative reference sequences of the egl gene of the sequevars described in the literature for the strains of Ralstonia spp. available in GenBank were inserted in the phylogenetic analysis to determine the sequevars. The bars indicate the fraction of substitutions per site. (TIF 161 kb)
Supplementary Fig. S4
Phylogenetic reconstruction based on partial mutS gene sequences from strains of Ralstonia pseudosolanacearum and R. solanacearum causing bacterial wilt in the semiarid climate of Pernambuco state, Brazil. This was conducted using the neighbor-joining method with the Jukes and Cantor algorithm and with 2000 bootstraps to test the tree topology in MEGA 7.0 software. Representative reference sequences of the mutS gene of the sequevars described in the literature for the strains of Ralstonia spp. available in GenBank were inserted in the phylogenetic analysis to determine the sequevars. The bars indicate the fraction of substitutions per site. (BMP 4007 kb)
ESM 5
(PNG 2.17 mb)
Supplementary Fig. S5
Phylogenetic reconstruction based on partial egl gene sequences from strains of Ralstonia pseudosolanacearum and R. solanacearum causing bacterial wilt in the semiarid climate of Pernambuco state, Brazil. This was conducted using maximum likelihood estimation in PhyML software version 3.1, and the TIM1 + G model was determined using JModel Test software v. 2.1.6. Representative reference sequences of the egl gene of the sequevars described in the literature for the strains of Ralstonia spp. available in GenBank were inserted in the phylogenetic analysis to determine the sequevars. The bars indicate the fraction of substitutions per site. (TIF 143 kb)
Supplementary Fig. S6
Phylogenetic reconstruction based on partial mutS gene sequences from strains of Ralstonia pseudosolanacearum and R. solanacearum causing bacterial wilt in the semiarid climate of Pernambuco state, Brazil. This was conducted using maximum likelihood estimation in PhyML software version 3.1, and the HKY + I model was determined using JModel Test software v. 2.1.6. Representative reference sequences of the mutS gene of the sequevars described in the literature for the strains of Ralstonia spp. available in GenBank were inserted in the phylogenetic analysis to determine the sequevars. The bars indicate the fraction of substitutions per site. (BMP 4530 kb)
Supplementary Table S1
(PDF 35 kb)
Supplementary Table S2
(XLSX 13.7 kb)
Supplementary Table S3
(XLSX 20 kb)
Supplementary Table S4
(PDF 34.7 kb)
Rights and permissions
About this article
Cite this article
Albuquerque, G.M.R., Silva, A.M.F., Silva, J.R. et al. Sequevar distribution of Ralstonia spp. in Solanaceae in the semiarid climate of the Pernambuco state, Brazil. Eur J Plant Pathol 159, 13–25 (2021). https://doi.org/10.1007/s10658-020-02132-4
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10658-020-02132-4