Abstract
The precipitous drop in the cost of genomic sequencing and the concomitant availability of computational methods for comparing genome-level data has made the accurate taxonomic placement of bacteria affordable and relatively rapid. Inaccurate taxonomic placement of bacteria has serious implications in clinical, environmental, and regulatory microbiology, but it can also adversely affect interpretation of research results. The quorum biosensor strain CV026 was derived from an isolate of Chromobacterium that was labeled as C. violaceum ATCC 31532, and is catalogued by the ATCC under that species name. Nearly 200 papers have been published that use CV026 as an indicator for quorum sensing activity in many Gram negative bacteria, but the inability of C. violaceum strains to complement the quorum sensing mutation in CV026 has called the taxonomic placement of the parent strain into question. We used molecular phylogeny and a large number of metabolic and phenotypic characters to demonstrate that Chromobacterium strain ATCC 31532 is a member of species Chromobacterium subtsugae.

Similar content being viewed by others
Key points
-
Chromobacterium strain CV026 is an important biosensor tool for quorum sensing research but its parent strain ATCC 31532 has been misidentified as C. violaceum.
-
Molecular phylogeny based on whole genome sequences and phenotypic characteristics indicate that Chromobacterium strain ATCC 31532 is most accurately placed in the species C. subtsugae.
-
Chromobacterium strain ATCC 31532 and C. subtsugae isolates do not produce HCN, but they do complement the CV026 quorum sensing cviI mutation.
Introduction
The discovery of quorum sensing (QS) as a means of intra- and extra-cellular bacterial communication in 1970 (at the time it was called ‘autoinduction’) (Nealson et al. 1970), and the recognition of QS as a general phenomenon (see for example Whitehead et al. 2001) has resulted in the development and availability of a number of QS biosensors that have been extremely valuable for the discovery and classification of the N-acyl homoserine lactones (AHLs) that serve as the primary autoinducers in Gram negative bacteria (Eberhard et al. 1981; McClean et al. 1997). Among the most commonly used biosensors is a mini-Tn5 cviI mutant of Chromobacterium strain ATCC 31532 known as CV026 (McClean et al. 1997; Chernin et al. 1998). Strain CV026 produces an easily-detectable purple phenotype due to the QS-dependent expression of the genes that encode the pigment violacein when complemented with an inducing concentration of medium chain-length HSLs (Chernin et al. 2011; McClean et al. 1997). As of the writing of this manuscript, there are 176 publications indexed in Pubmed since 1997 that use CV026 as a biosensor. However, there has been confusion about the correct taxonomic placement of Chromobacterium strain ATCC 31532, which is listed by the ATCC and other national culture collections as Chromobacterium violaceum, the genomic sequence of a Tn5 mutant of strain ATCC 31532 is catalogued in the GenBank database as C. subtsugae CV017 (accession number LKIW01000000), and the vast majority of publications using CV026 refer to it as C. violaceum. Because interpretation of QS complementation data can be affected by the taxonomic placement of the strain, it is important to correct the provenance of the parent strain and its QS mutant as an important tool in microbiology.
The species Chromobacterium violaceum was first proposed by Bergonzini in 1881 (Gillis and Logan 2015), and ATCC 12472 is the type strain of the species (syn. NCIB 9131, NCTC 9757). In 1982, strain ATCC 31532 was published and deposited in the American Type Culture Collection (ATCC) as C. violaceum due to similar phenotypic characteristics associated with C. violaceum ATCC 12472T (Wells et al. 1982). C. subtsugae was characterized and added to the genus in 2007 (Martin et al. 2007). The genus Chromobacterium has further expanded since then with nearly a dozen new species published within the last decade. The absence of affordable and available genomic sequencing tools until the last few years, and the rapid increase in the number of recognized species may have contributed to errors in Chromobacterium spp. identification (Kampfer et al. 2018). A recently published study on the reclassification of C. pseudoviolaceum LMG3953 showed genomic evidence that Chromobacterium strain ATCC 31532 and type strain C. subtsugae PRAA4-1T (Martin et al. 2007) were similar enough to warrant a reclassification of strain ATCC 31532 (Kampfer et al. 2018).
In this work we provide genotypic and phenotypic evidence that strain ATCC 31532 is clearly a member of the species C. subtsugae.
Materials and methods
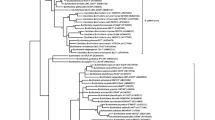
To correctly place strain ATCC 31532 within the genus, we constructed a Chromobacterium spp. phylogenetic tree using TYGS (https://tygs.dsmz.de/) with default settings to confirm that Chromobacterium strain ATCC 31532 clusters with C. subtsugae PRAA4-1T rather than with C. violaceum ATCC 12472T the type isolate of the original identification (Fig. 1). This tree is based on the Genome BLAST Distance Phylogeny approach using FastME (Meier-Kolthoff et al. 2019). A second tree was generated with the Codon Trees pipeline in PATRIC version 3.5.43 (https://www.patricbrc.org/) using default settings (Wattam et al. 2017) (Fig. 2) with all of the currently-available Chromobacterium spp. genome sequences. Amino acid and nucleotide sequences from a defined number of PGFams (Davis et al. 2016) were randomly selected from a list of single copy genes to build an alignment. A tree was then generated from the differences within the sequences. Protein sequences were aligned using MUSCLE (Edgar et al. 2004), and nucleotide coding gene sequences were aligned using the Codon_align function of BioPython (Cock et al. 2009). No deletions or duplications were allowed in the alignment. One hundred of a possible 423 single-copy coding sequences were used, resulting in alignment of 45,762 amino acids and 137,286 nucleotides. A concatenated alignment of all proteins and nucleotides was used to generate a partitions file for RAxML version 8.2.11 (Stamatakis et al. 2014). Support values were calculated using 100 rounds of the ‘Rapid’ bootstrapping function of RAxML (Stamatakis et al. 2008). Twenty-two Chromobacterium spp. genomic sequences were used to compile the tree from genomic sequences of the family Chromobacteriaceae with the β-proteobacteria Aquitalea magnusonii and Bordetella bronchiseptica as outgroups. Additional overall genome relatedness indices (OGRI) comparisons based on variations of Average Nucleotide Identity (ANIb and orthoANIu) were performed using online tools available through JSpeciesWS (http://jspecies.ribohost.com/jspeciesws/) (Richter et al. 2015), digital DNA-DNA hybridization (dDDH) with TYGS and GGDC 2.1 (http://ggdc.dsmz.de/) (Meier-Kolthoff et al. 2013), and EZBioCloud (ezhttps://www.ezbiocloud.net/tools/ani) (Yoon et al. 2017) (Table 1).
A RAxML phylogenetic tree was constructed in PATRIC using 100 aligned single copy amino acid and nucleotide sequences from a defined number of PGFams. Only bootstrap values less than 100 are shown at the nodes. Accession numbers for genome sequences are shown in parentheses. The isolate known as C. violaceum ATCC315322 is unambiguously located in the C. subtsugae clade, rather than the C. violaceum clade. The β-proteobacteria Aquitalea magnusonii and Bordetella bronchiseptica serve as outgroups for the family and phylum. The tree is rooted on the B. bronchiseptica branch. The scale bar equals 0.02 substitutions
Phenotypic and metabolic tests comparing isolates C. violaceum ATCC 12472T, C. subtsugae ATCC 31532 and C. subtsugae PRAA4-1T were conducted using API 20 NE test strips (BioMerieux) and Biolog GEN III plates according to the manufacturers’ instructions. Altogether we generated comparative data from over 100 different metabolic and phenotypic tests. Production of hydrogen cyanide (HCN), and AHL complementation tests were also conducted to further increase the reliability of taxonomic assignment of strain ATCC 31532 to either the C. violaceum or the C. subtsugae subclades. HCN quantification was performed as previously reported (Ebadzadsahrai et al. 2020). Cells were grown to late log phase at 26 °C in King’s Medium B broth and pelleted by centrifugation. Cell-free supernatant pH was adjusted to pH 11 by the addition of 50 µl 1N NaOH per ml supernatant, and measured directly using an ion-specific electrode (Lazar Research Laboratories) as previously described (Zlosnik and Williams 2004). Measurements were converted to ppm with a standard curve.
AHL complementation of the QS mutant CV026 was determined by growing the biosensor strain adjacent to strain ATCC 31532, C. subtsugae PRAA4-1T, or C. violaceum ATCC 12472T on King’s Medium B agar plates for 24 h at room temperature. The biosensor strain does not produce its own AHL, therefore it does not produce the purple pigment violacein. In this assay, complementation of the cviI mutation occurs by diffusion of exogenously applied or biologically produced AHLs through the agar to the biosensor strain in sufficient concentration to trigger the QS response, resulting in the production of violacein.
Results
Phylogenetic evidence
Strain ATCC 31532 is located within the C. subtsugae clade of both GDBP and RAxML Chromobacterium spp. genomic phylogenetic trees, indicating its close relationship to that species, and not within the C. violaceum or other clades (Figs. 1 and 2). All of the other clades were internally consistent, and had strong bootstrap support in both trees. OGRI comparisons using three different algorithms, ANIb, OrthoANIu, and dDDH confirm the close relatedness of Chromobacterium strain ATCC 31532 to C. subtsugae, particularly to strain F49, but also well within the defining limits of 95–96% for ANI, 70% for dDDH, and 1% for G+C with all of the C. subtsugae strains in our collection, and well outside the limits of C. violaceum ATCC 12472 and the other Chromobacterium species (Table 1).
Phenotypic evidence
Most metabolic and phenotypic characteristics examined were shared among all three isolates, as would be expected for members of the same genus, but 22 characteristics were the same for strain ATCC 31532 and C. subtsugae PRAA4-1T that differed from C. violaceum ATCC 12472T (Table 2). There were no metabolic differences between strain ATCC 31532 and C. subtsugae PRAA4-1T. The type isolate C. violaceum ATCC 12472 reduced nitrate, hydrolyzed aesculin, and utilized d-mannose, sucrose, adipic acid, pectin, and glucuronamide whereas strain ATCC 31532 and TC. subtsugae PRAA4-1T did not. Strain ATCC 31532 and C. subtsugae PRAA4-1T utilized dextrin, a-d-lactose, d-salicin, d-galactose, 3-methyl glucose, l-fucose, d-arabitol, myo-inositol, l-pyroglutamic acid, citrc acid, and N-acetyl-d-galactosamine, whereas C. violaceum ATCC 12472T did not. HCN is produced by both C. violaceum (Michaels and Corpe 1965) and C. vaccinii (Voing et al. 2015; Ebadzadsahrai et al. 2020), but has not been reported from C. subtsugae. C. violaceum ATCC 12472T supernatants contained 81 ppm HCN, C. subtsugae PRAA4-1T, MWU 12-2387, and strain ATCC 31532 supernatants contained about 10 ppm, which is the baseline level of the growth medium (Fig. 2). CV026, the cviI mutant of strain ATCC 31532, was complemented by its parent strain ATCC 31532 as well as by C. subtsugae PRAA4-1T, and C. subtsugae MWU 12-2387 (Voing et al. 2017), but not by C. violaceum ATCC 12472T, which indicates that strain ATCC 31532, C. subtsugae PRAA4-1T, and C. subtsugae MWU 12-2387 produce N-acyl-homoserine lactones (AHLs) that are recognized by CV026, but the AHLs produced by C. violaceum ATCC 12472 T are not.
Discussion
Taxonomic placement of Chromobacterium strain ATCC 31532, the parent strain of the widely-used QS biosensor CV026, is critical for the correct interpretation of complementation experiments because C. violaceum and C. subtsugae use different autoinducers (Stauff and Bassler 2011; Rekha et al. 2011; McClean et al. 1997). We have used both genotypic and phenotypic evidence to show that the parent strain of the biosensor CV026 has been misidentified and deposited in the American Type Tissue Collection (ATCC) and other national repositories as C. violaceum because it was deposited long before C. subtsugae had been described and included in validated prokaryote species lists. Several complementary phylogenetic and genotypic analyses confirm the placement of Chromobacterium strain ATCC 31532 in the species C. subtsugae, including very different types of OGRI algorithms (Table 1). A large number of metabolic and other phenotypic tests also indicate that strain ATCC 31532 is a member of the species C. subtsugae, and not C. violaceum (Table 2). The primary AHL produced by C. subtsugae PRAA4-1T is N-hexanoyl-l-homoserine lactone (C6-HSL) (McClean 1997) whereas the AHL produced by C. violacein ATCC 12472T is N-decanoyl-l-homoserine lactone (C10-HSL) (Morohoshi et al. 2008). Further evidence is the production of HCN by C. violaceum but not by C. subtsugae strains, including ATCC 31532 (Fig. 3). Based on both genotypic and phenotypic characterization, it is clear that Chromobacterium strain ATCC 31532, and therefore the widely used biosensor strain CV026, have been misidentified as C. violaceum, and should be re-designated C. subtsugae ATCC 31532.
Availability of data and materials
All data used in the phylogenetic analysis are publicly available in GenBank. Microbial strains are publicly available, or in the case of C. subtsugae lab strains may be requested from the authors.
Code availability
Not applicable.
References
Chernin LS, Winson MK, Thompson JM, Haran S, Bycroft BW, Chet I, Williams P, Stewart G (1998) Chitinolytic activity in Chromobacterium violaceum: substrate analysis and regulation by quorum sensing. J Bacteriol 180(17):4435–4441
Chernin L, Toklikishvili N, Ovadis M, Kim S, Ben-Ari J, Khmel I, Vainstein A (2011) Quorum-sensing quenching by rhizobacterial volatiles. Environ Microbiol Rep 3(6):698–704. doi:https://doi.org/10.1111/j.1758-2229.2011.00284.x
Cock PJ, Antao T, Chang JT, Chapman BA, Cox CJ, Dalke A, Friedberg I, Hamelryck T, Kauff F, Wilczynski B, de Hoon MJ (2009) Biopython: freely available Python tools for computational molecular biology and bioinformatics. Bioinformatics 25(11):1422–1423. doi:https://doi.org/10.1093/bioinformatics/btp163
Davis JJ, Gerdes S, Olsen GJ, Olson R, Pusch GD, Shukla M, Vonstein V, Wattam AR, Yoo H (2016) PATtyFams: Protein families for the microbial genomes in the PATRIC database. Front Microbiol 7:118. https://doi.org/10.3389/fmicb.2016.00118
Ebadzadsahrai G, Higgins Keppler EA, Soby SD, Bean HD (2020) Inhibition of fungal growth and induction of a novel volatilome in response to Chromobacterium vaccinii volatile organic compounds. Front Microbiol. https://doi.org/10.3389/fmicb.2020.01035
Eberhard A, Burlingame AL, Eberhard C, Kenyon GL, Nealson KH, Oppenheimer NJ (1981) Structural identification of autoinducer of Photobacterium fischeri luciferase. Biochemistry 20(9):2444–2449. doi:https://doi.org/10.1021/bi00512a013
Edgar RC (2004) MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res 32(5):1792–1797. doi:https://doi.org/10.1093/nar/gkh340
Gillis M, Logan NA (2015) Chromobacterium. Bergey’s Manual of Systematics of Archaea and Bacteria. Wiley, Hoboken. https://doi.org/10.1002/9781118960608.gbm00975
Kampfer P, Glaeser SP, Soby SD et al (2018) Chromobacterium pseudoviolaceum Kampfer et al. 2009 is a later heterotypic synonym of Chromobacterium violaceum Bergonzini 1880. Int J Syst Evol Microbiol 68(9):2967–2968. https://doi.org/10.1099/ijsem.0.002930
Martin PA, Gundersen-Rindal D, Blackburn M, Buyer J (2007) Chromobacterium subtsugae sp. nov., a betaproteobacterium toxic to Colorado potato beetle and other insect pests. Int J Syst Evol Microbiol 57(Pt 5):993–999. doi:https://doi.org/10.1099/ijs.0.64611-0
McClean KH, Winson MK, Fish L, Taylor A, Chhabra SR, Camara M, Daykin M, Lamb JH, Swift S, Bycroft BW, Stewart GS, Williams P (1997) Quorum sensing and Chromobacterium violaceum: exploitation of violacein production and inhibition for the detection of N-acylhomoserine lactones. Microbiology 143(Pt 12):3703–3711
Meier-Kolthoff JP, Göker M (2019) TYGS is an automated high-throughput platform for state-of-the-art genome-based taxonomy. Nat Commun 10:2182. https://doi.org/10.1038/s41467-019-10210-3
Meier-Kolthoff JP, Auch AF, Klenk H-P, Göker M (2013) Genome sequence-based species delimitation with confidence intervals and improved distance functions. BMC Bioinformatics 14:60
Michaels R, Corpe WA (1965) Cyanide formation by Chromobacterium violaceum. J Bacteriol 89:106–112
Morohoshi T, Kato M, Fukamachi K, Kato N, Ikeda T (2008) N-acylhomoserine lactone regulates violacein production in Chromobacterium violaceum type strain ATCC 12472. FEMS Microbiol Lett 279(1):124–130. doi:https://doi.org/10.1111/j.1574-6968.2007.01016
Nealson KH, Platt T, Hastings JW (1970) Cellular control of the synthesis and activity of the bacterial luminescent system. J Bacteriol 104(1):313–322
Rekha PD, Young CC, Arun AB (2011) Identification of N-Acyl-L-homoserine lactones produced by non-pigmented Chromobacterium aquaticum CC-SEYA-1 and pigmented Chromobacterium subtsugae PRAA4-1. 3 Biotech 1(4):1–7. doi:https://doi.org/10.1007/s13205-011-0029-1
Richter M, Rosselló-Móra R, Glöckner FO, Peplies J (2015) JSpeciesWS: a web server for prokaryotic species circumscription based on pairwise genome comparison. Bioinformatics. 32(6):929
Stamatakis A (2014) RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics 30(9):1312–1313. doi:https://doi.org/10.1093/bioinformatics/btu033
Stamatakis A, Hoover P, Rougemont J (2008) A rapid bootstrap algorithm for the RAxML web ssrvers. Syst Biol 57(5):758–771. doi:https://doi.org/10.1080/10635150802429642
Stauff DL, Bassler BL (2011) Quorum sensing in Chromobacterium violaceum: DNA recognition and gene regulation by the CviR receptor. J Bacteriol 193(15):3871–3878. doi:https://doi.org/10.1128/JB.05125-11
Voing K, Harrison A, Soby SD (2015) Draft genome sequence of Chromobacterium vaccinii, a Potential Biocontrol Agent against Mosquito (Aedes aegypti) larvae. Genome Announc 3(3):e00477–e00415. doi:https://doi.org/10.1128/genomeA.00477-15
Voing K, Harrison A, Soby SD (2017) Draft genome sequence of Chromobacterium subtsugae MWU12-2387 isolated from a wild cranberry bog in Truro. Genome Announc, Massachusetts. https://doi.org/10.1128/genomeA.01633-16
Wattam AR, Davis JJ, Assaf R, Boisvert S, Brettin T, Bun C, Conrad N, Dietrich EM, Disz T, Gabbard JL, Gerdes S, Henry CS, Kenyon RW, Machi D, Mao C, Nordberg EK, Olsen GJ, Murphy-Olson DE, Olson R, Overbeek R, Parrello B, Pusch GD, Shukla M, Vonstein V, Warren A, Xia F, Yoo H, Stevens RL (2017) Improvements to PATRIC, the all-bacterial Bioinformatics Database and Analysis Resource Center. Nucleic Acids Res 45(D1):D535–D542. doi:https://doi.org/10.1093/nar/gkw1017
Wells JS, Trejo WH, Principe PA, Bush K, Georgopapadakou N, Bonner DP, Sykes RB (1982) SQ 26,180, a novel monobactam. I Taxonomy, fermentation and biological properties. J Antibiot (Tokyo) 35(2):184–188. doi:https://doi.org/10.7164/antibiotics.35.184
Whitehead NA, Barnard AML, Slater H, Simpson NJL, Salmond GPC (2001) Quorum-sensing in gram-negative bacteria. FEMS Microbiol Rev 25:365–404
Yoon SH, Ha SM, Lim JM, Kwon SJ, Chun J (2017) A large-scale evaluation of algorithms to calculate average nucleotide identity. Antonie van Leeuwenhoek 110:1281–1286
Zlosnik JEA, Williams HD (2004) Methods for assaying cyanide in bacterial culture supernatant. Lett Appl Microbiol 38(5):360–365. doi:https://doi.org/10.1111/j.1472-765X.2004.01489
Acknowledgements
This work was funded internally by the Biomedical Sciences Program, College of Graduate Studies, Midwestern University.
Funding
This research was supported by Midwestern University intramural funds.
Author information
Authors and Affiliations
Contributions
AH and SS conceived and designed the research, and wrote the manuscript. AH conducted experiments and compiled the data. SS analyzed the data. Both authors read and approved the final manuscript.
Corresponding author
Ethics declarations
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no conflicting or competing interests with the work presented.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Harrison, A.M., Soby, S.D. Reclassification of Chromobacterium violaceum ATCC 31532 and its quorum biosensor mutant CV026 to Chromobacterium subtsugae. AMB Expr 10, 202 (2020). https://doi.org/10.1186/s13568-020-01140-1
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s13568-020-01140-1