Abstract
Eight novel strains of the phylum Planctomycetes were isolated from different aquatic habitats. Among these habitats were the hydrothermal vent system close to Panarea Island, a public beach at Mallorca Island, the shore of Costa Brava (Spain), and three sites with brackish water in the Baltic Sea. The genome sizes of the novel strains range from 4.33 to 6.29 Mb with DNA G+C contents between 52.8 and 66.7%. All strains are mesophilic (Topt 24–30 °C) and display generation times between 17 and 94 h. All eight isolates constitute novel species of either already described or novel genera within the family Lacipirellulaceae. Two of the novel species, Posidoniimonas polymericola (type strain Pla123aT = DSM 103020T = LMG 29466T) and Bythopirellula polymerisocia (type strain Pla144T = DSM 104841T = VKM B-3442T), belong to established genera, while the other strains represent the novel genera Aeoliella gen. nov., Botrimarina gen. nov., Pirellulimonas gen. nov. and Pseudobythopirellula gen. nov. Based on our polyphasic analysis, we propose the species Aeoliella mucimassa sp. nov. (type strain Pan181T = DSM 29370T = LMG 31346T = CECT 9840T = VKM B-3426T), Botrimarina colliarenosi sp. nov. (type strain Pla108T = DSM 103355T = LMG 29803T), Botrimarina hoheduenensis sp. nov. (type strain Pla111T = DSM 103485T = STH00945T, Jena Microbial Resource Collection JMRC), Botrimarina mediterranea sp. nov. (type strain Spa11T = DSM 100745T = LMG 31350T = CECT 9852T = VKM B-3431T), Pirellulimonas nuda sp. nov. (type strain Pla175T = DSM 109594T = CECT 9871T = VKM B-3448T) and Pseudobythopirellula maris sp. nov. (type strain Mal64T = DSM 100832T = LMG 29020T).
Similar content being viewed by others
Introduction
Together with Verrucomicrobia, Chlamydiae and other sister phyla, the bacterial phylum Planctomycetes is part of the PVC superphylum (Wagner and Horn 2006). According to the current taxonomy, the phylum Planctomycetes comprises the phylogenetic classes Planctomycetia, Phycisphaerae and Candidatus Brocadiae. Members of the class Planctomycetia have been found to occur ubiquitously but are predominant in aquatic environments (Wiegand et al. 2018). They have been isolated from numerous algal surfaces (Bengtsson et al. 2012; Boersma et al. 2019; Bondoso et al. 2014, 2015, 2017; Kallscheuer et al. 2019a, d; Lage and Bondoso 2014; Peeters et al. 2019; Vollmers et al. 2017; Waqqas et al. 2020), on which they sometimes even dominate the microbial community (Bengtsson and Øvreås 2010; Wiegand et al. 2018). Planctomycetes are considered important players in global carbon cycling (Wiegand et al. 2018) as they possess the metabolic ability to degrade complex carbon substrates (Jeske et al. 2013; Lachnit et al. 2013). Planctomycetes are suspected to produce small bioactive molecules (Graca et al. 2016; Jeske et al. 2016; Waqqas et al. 2020). Recently, stieleriacines, one of the first small molecules elucidated from planctomycetes, were shown to be involved in alteration of community compositions in biofilms (Kallscheuer et al. 2020a). This finding might lead to an explanation as to why the cells can dominate algal surfaces without being outcompeted, despite their slow growth rates (Wiegand et al. 2018).
Members of the class Planctomycetia divide by budding, without employing otherwise essential divisome proteins such as the canonically used division ring-assembling FtsZ (Jogler et al. 2012; Kallscheuer et al. 2019d; Pilhofer et al. 2008). Most of the species seem to perform a lifestyle switch during their development, alternating between planktonic, motile swimmer cells and sessile mother cells (Gade et al. 2005). Their periplasm can be extremely enlarged and compartmentalised (Acehan et al. 2013; Boedeker et al. 2017), most likely to enable the digestion of internalised polysaccharides (Boedeker et al. 2017). Interestingly, Planctomycetes were only recently found to possess a peptidoglycan cell wall (Jeske et al. 2015; van Teeseling et al. 2015), which was also found in the closely related Verrucomicrobia (Rast et al. 2017).
The class Planctomycetia was recently reclassified and now comprises four different orders: Pirellulales, Gemmatales, Isosphaerales and the revised Planctomycetales (Dedysh et al. 2020). The latter three taxa harbour only a single family each, whereas the order Pirellulales currently consists of three families: Pirellulaceae, which contain most described genera and species (Kallscheuer et al. 2019b, c, d, 2020c; Rensink et al. 2020; Schubert et al. 2020); Thermoguttaceae, which feature the only known thermophilic Planctomycetes (Slobodkina et al. 2015, 2016); and Lacipirellulaceae (Dedysh et al. 2020). In addition to the type species of the type genus, Lacipirellula parvula, other described members of the family are ‘Lacipirellula limnantha’ (Kallscheuer et al. in revision), ‘Bythopirellula goksoyri’ (Storesund and Øvreås 2013) and Posidoniimonas corsicana (Kohn et al. 2020a).
In this study, we extend the current collection of Lacipirellulaceae strains by the description of eight novel species belonging to two known and four hitherto undescribed genera.
Materials and methods
Isolation of the novel strains and cultivation conditions
For strain isolation and cultivation, M1H NAG ASW medium was used. For medium preparation, 0.25 g peptone (Bacto), 0.25 g yeast extract (Bacto), 2.38 g (4-(2-hydroxyethyl)-1-piperazineethane-sulfonic acid) (HEPES) (10 mM), 250 mL artificial seawater (ASW) and 20 mL Hutner’s basal salt solution were mixed in a final volume of 973 mL double distilled water. The pH was adjusted to 7.5 using 5 M KOH and the solution was autoclaved for 20 min at 121 °C. After cooling, the following solutions were added aseptically: 1 mL of 25% (w/v) glucose, 5 mL vitamin solution, 1 mL trace element solution and 20 mL of a stock solution with 50 g/L N-acetyl glucosamine (NAG). ASW contained 46.94 g/L NaCl, 7.84 g/L Na2SO4, 21.28 g/L MgCl2·6 H2O, 2.86 g/L CaCl2·2 H2O, 0.384 g/L NaHCO3, 1.384 g/L KCl, 0.192 g/L KBr, 0.052 g/L H3BO3, 0.08 g/L SrCl2·6 H2O and 0.006 g/L NaF and was freshly prepared before addition to the base solution. Hutner’s basal salt solution was prepared by first dissolving 10 g nitrilotriacetic acid (NTA) in 700 mL double distilled water and adjusting the pH to 7.2 using 5 M KOH. Subsequently, the following compounds were added: 29.7 g MgSO4·7 H2O, 3.34 g CaCl2·2 H2O, 0.01267 g Na2MoO4·2 H2O, 0.099 g FeSO4·7 H2O and 50 mL metal salt solution 44. The solution was filled up to 1 L, sterilised by filtration and stored at 4 °C. Metal salt solution 44 consisted of 250 mg/L Na2-EDTA, 1095 mg/L ZnSO4·7 H2O, 500 mg/L FeSO4·7 H2O, 154 mg/L MnSO4·H2O, 39.5 mg/L CuSO4·5 H2O, 20.3 mg/L CoCl2·6 H2O and 17.7 mg/L Na2B4O7·10 H2O. In the first step, EDTA was dissolved and, if required, a few drops of concentrated H2SO4 were added to retard precipitation of the heavy metal ions. Metal salt solution 44 was sterilised by filtration and stored at 4 °C. Vitamin solution contained per litre: 10 mg p-aminobenzoic acid, 4 mg biotin, 20 mg pyridoxine hydrochloride, 10 mg thiamine hydrochloride, 10 mg calcium pantothenate, 4 mg folic acid, 10 mg riboflavin, 10 mg nicotinamide and 0.2 mg vitamin B12. p-Aminobenzoic acid was dissolved first and the solution was sterilised by filtration and stored in the dark at 4 °C. The trace element solution containing 1.5 g/L Na-nitrilotriacetate, 500 mg/L MnSO4·H2O, 100 mg/L FeSO4·7 H2O, 100 mg/L Co(NO3)2·6 H2O, 100 mg/L ZnCl2, 50 mg/L NiCl2·6 H2O, 50 mg/L H2SeO3, 10 mg/L CuSO4·5 H2O, 10 mg/L AlK(SO4)2·12 H2O, 10 mg/L H3BO3, 10 mg/L NaMoO4·2 H2O and 10 mg/L Na2WO4·2 H2O was sterilised by filtration and stored in the dark at 4 °C. The sampling locations and initial isolation of all strains are listed in Table S1. Sampled material was swabbed over or streaked on plates containing M1H NAG ASW medium solidified with 8 g/L gellan gum and additionally supplemented with 500 mg/L streptomycin, 200 mg/L ampicillin and 20 mg/L cycloheximide. The use of the two antibiotics and the anti-fungal agent cycloheximide is part of the enrichment strategy targeting strains of the phylum Planctomycetes, which are known to be resistant to both antibiotics. Colonies obtained after the first round of cultivation (four to six weeks) were streaked on a new plate and subsequently maintained in liquid M1H NAG ASW medium. Initial amplification and sequencing of the 16S rRNA gene was performed as previously described (Rast et al. 2017). This step was included to ensure that strains chosen for detailed characterisation are indeed members of the phylum Planctomycetes.
Light microscopy
Phase contrast light microscopic analyses were performed employing a Nikon Eclipse Ti inverted microscope with a Nikon DS-Ri2 camera (blue LED). Specimens were immobilised in MatTek glass bottom dishes (35 mm, No. 1.5) employing a 1% (w/v) agarose cushion (Will et al. 2018). Images were analysed using the Nikon NIS-Elements software (version 4.3). To determine the cell size, at least 100 representative cells were counted manually (Annotations and Measurements, NIS-Elements) or by using the NIS-Elements semi-automated object count tool (smooth: 4×, clean: 4×, fill holes: on, separate: 4×). The Object Count tool enables setting a threshold for the image, automatically measures the binary objects and exports the measured data to a file.
Electron microscopy
For field emission scanning electron microscopy, bacteria were fixed in 1% (v/v) formaldehyde in HEPES buffer (3 mM HEPES, 0.3 mM CaCl2, 0.3 mM MgCl2, 2.7 mM sucrose, pH 6.9) for 1 h on ice and washed once employing the same buffer (Rast et al. 2017). Cover slips with a diameter of 12 mm were coated with a poly-l-lysine solution (Sigma-Aldrich) for 10 min, washed in distilled water and air-dried. 50 µL of the fixed bacteria solution was placed on a cover slip and allowed to settle for 10 min. Cover slips were then fixed in 1% (v/v) glutaraldehyde in TE buffer (20 mM TRIS, 1 mM EDTA, pH 6.9) for 5 min at room temperature and subsequently washed twice with TE buffer before dehydrating in a graded series of acetone (10, 30, 50, 70, 90, 100%) on ice for 10 min. at each concentration. Samples from the 100% acetone step were brought to room temperature before placing them in fresh 100% acetone. Samples were then subjected to critical-point drying with liquid CO2 (CPD 300, Leica). Dried samples were covered with a gold/palladium (80/20) film by sputter coating (SCD 500, Bal-Tec) before examination in a field emission scanning electron microscope (Zeiss Merlin) using the Everhart–Thornley HESE2 detector and the in-lens SE detector in a 25:75 ratio at an acceleration voltage of 5 kV.
Physiological analyses
For determination of the temperature optimum for growth, all strains were cultivated in M1H NAG ASW medium at pH 7.5 at different temperatures ranging from 10 to 40 °C. For determination of the pH optimum for growth, 100 mM HEPES was used instead of 10 mM for cultivations at pH 7.0, 7.5 and 8.0. For cultivation at pH 5.0, 5.5 and 6.0 HEPES was replaced by 100 mM 2-(N-morpholino)ethanesulfonic acid (MES), whereas 100 mM 3-(4-(2-hydroxyethyl)piperazin-1-yl)propane-1-sulfonic acid (HEPPS) was used as buffering agent at pH 8.5 and 100 mM N-cyclohexyl-2-aminoethanesulfonic acid (CHES) at pH 9.0, 9.5 and and 10.0. Cultivations for determination of the pH optimum were performed at 28 °C. Cell densities were determined as optical density at 600 nm (OD600).
Genome information and genome-based analyses
The genomes of all strains were published previously (Wiegand et al. 2020) and are available from RefSeq under accession numbers SJPQ00000000 (strain Mal64T), CP036278 (strain Pan181T), SJPR00000000 (strain Pla108T), SJPH00000000 (strain Pla111T), SJPO00000000 (Pla123aT), SJPS00000000 (Pla144T), CP036291 (strain Pla175T) and CP036349 (strain Spa11T). The GenBank accession numbers of the respective 16S rRNA genes are MK554544 (strain Mal64T), MK559982 (strain Pan181T), MK554547 (Pla108T), MK554579 (strain Pla111T), MK554580 (strain Pla123aT), MK554548 (strain Pla144T), MK559987 (strain Pla175T) and MK554534 (strain Spa11T).
A genome-based analysis of enzymes participating in central carbon metabolism was conducted by examining locally computed InterProScan (Mitchell et al. 2019) results cross-referenced with information from the UniProt (UniProt 2019) database and BLASTp results of typical protein sequences. The analysis of the pan genomes was performed with anvi’o (Eren et al. 2015), following the pangenomics workflow (Delmont and Eren 2018).
Construction of phylogenetic trees for the novel isolates
Maximum likelihood 16S rRNA gene sequence-based phylogeny was computed for the eight novel strains, the type strains of all recently described planctomycetal species (as of June 2020) (Kallscheuer et al. 2020b; Kumar et al. 2020; Peeters et al. 2020; Rivas-Marin et al. 2020a, b; Schubert et al. 2020) and all isolates recently published, but not yet described (Wiegand et al. 2020). The 16S rRNA gene sequences were aligned with SINA (Pruesse et al. 2012). The phylogenetic analysis was performed employing a maximum likelihood approach with 1000 bootstraps, the nucleotide substitution model GTR, gamma distribution and estimation of proportion of invariable sites (GTRGAMMAI option) (Stamatakis 2014). Three 16S rRNA genes of bacterial strains from the PVC superphylum (Lentisphaera araneosa, acc. no. ABCK01; Kiritimatiella glycovorans, acc. no. CP010904.1 and Opitutus terrae, acc. no. AJ229235.1) served as outgroup.
For the multilocus sequence analysis (MLSA) the unique single-copy core genome of all analysed genomes was determined with proteinortho5 (Lechner et al. 2011) with the ‘selfblast’ option enabled. The protein sequences of the resulting orthologous groups were aligned using MUSCLE v.3.8.31 (Edgar 2004). After clipping, partially aligned C- and N-terminal regions and poorly aligned internal regions were filtered using Gblocks (Castresana 2000). The final alignment of 900 ubiquitous genes with a combined length of 440,140 conserved amino acid residues was concatenated and clustered using FastTree (Price et al. 2010). The outgroup consisted of three genomes from strains of the families Pirellulaceae and Planctomycetaceae (Bremerella volcania, acc. no. CP036289.1; Rubinisphaera brasiliensis, acc. no. CP002546.1 and Gimesia maris, acc. no. CP042910.1).
Analysis of phylogenetic markers
The rpoB nucleotide sequences (encoding the RNA polymerase β-subunit) were taken from publicly available genome annotations and the sequence identities were determined as described previously (Bondoso et al. 2013) using Clustal Omega (Sievers et al. 2011). Alignment and matrix calculation were performed only using those parts of the sequence that would have been sequenced with the previously described primer set (Bondoso et al. 2013). The average nucleotide identity (ANI) was calculated using OrthoANI (Lee et al. 2016). The average amino acid identity (AAI) was obtained with the aai.rb script of the enveomics collection (Rodriguez-R and Konstantinidis 2016) and the percentage of conserved proteins (POCP) was calculated as described before (Qin et al. 2014).
Results and discussion
Sampling at different locations in the Baltic Sea and the Mediterranean Sea
The strains Spa11T, Pla108T, Pla111T, Mal64T, Pla123aT, Pla175T, Pan181T and Pla144T were recently reported as novel isolates obtained during a large diversity-driven cultivation campaign targeting the phylum Planctomycetes (Wiegand et al. 2020). The eight strains were isolated from water or biofilms obtained from different aquatic sampling locations: a hydrothermal vent area offshore of Panarea island (Pan181T), phytoplankton from a public beach at Mallorca island (Mal64T) and seawater from the Costa Brava, Spain (Spa11T). Additionally, strains were recovered from wood or polyethylene particles incubated for two weeks at three different samplings spots with brackish water in or adjacent to the Baltic Sea (Pla108T, Pla111T, Pla123aT, Pla175T and Pla144T). More details regarding the sampling spots and the isolation are provided in Table S1.
Phylogeny
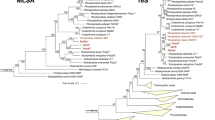
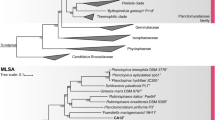
Both 16S rRNA gene sequence as well as the MLSA-based phylogenetic trees suggest that all the strains described here are members of the recently established planctomycetal family Lacipirellulaceae, order Pirellulales, class Planctomycetia, phylum Planctomycetes (Dedysh et al. 2020) (Fig. 1). By closer examination of the phylogenetic trees, these strains and reference strains can be subdivided in seven groups: (I) strains Spa11T, Pla108T and Pla111T; (II) P. corsicana KOR34T (Kohn et al. 2020a) and strain Pla123aT; (III) strain Pan181T; (IV) L. parvula PX69T (Dedysh et al. 2020) and ‘L. limnantha’ I41T (Kallscheuer et al. in revision); (V) strain Pla175T; (VI) ‘B. goksoyri’ Pr1dT (Storesund and Øvreås 2013) and strain Pla144T, and (VII) strain Mal64T.
Phylogeny of the class Planctomycetia based on 16S rRNA gene sequence and of the family Lacipirellulaceae using whole genome-based multilocus sequence analysis (MLSA). Included in both analyses are the strains described here, ‘Bythopirellula goksoyri’ (Storesund and Øvreås 2013), the recently described Lacipirellula parvula (Dedysh et al. 2020), ‘L. limnantha’ (Kallscheuer et al. in revision) and Posidoniimonas corsicana (Kohn et al. 2020a). For the 16S rRNA gene sequence analysis, maximum likelihood estimation-gained bootstrap values after 1000 re-samplings are given at the nodes in %. The outgroup consists of three 16S rRNA genes from the PVC superphylum outside of the phylum Planctomycetes. Members of the other classes in the phylum Planctomycetes are not shown in the tree. For the MLSA-based phylogeny, reliability estimators based on Shimodaira-Hasegawa testing were determined. As they were always 1, they are not shown in the tree. The outgroup contains three genomes from the family Planctomycetaceae
Evaluation of ANI indicated that all the novel strains belong to separate species, given that all values (< 80%) are clearly below the threshold of 95% for delineation of prokaryotic species (Kim et al. 2014) (Fig. 2 and Table S2). This is also supported by identity of a 1200 bp partial sequence of the rpoB gene, for which the species threshold was proposed to be between 95.5 and 98.2% (Bondoso et al. 2013). For this phylogenetic marker, all strains turned out to have minimal identity values of < 93.7% (Fig. 2 and Table S3). For 16S rRNA gene sequence identities the situation is less clear, as some values are above the well-established species threshold of 98.7% (Yarza et al. 2014) (Fig. 2 and Table S4). This is a phenomenon known for the families Planctomycetaceae and Pirellulaceae, in which some novel species have been found to have a 16S rRNA gene sequence identity of up to 99.9% (Kohn et al. 2020b; Rensink et al. 2020). The subsequent analysis of the markers AAI and POCP indicated that the different groups (I-VII) belong to different genera within the family Lacipirellulaceae. With the exception of group I vs. VII (AAI: 60.4%; POCP: 57.6%) all minimal AAI values are below the proposed genus threshold of 60–80% (Luo et al. 2014) and POCP values are very close (< 50.7%) or below the genus threshold of 50% (Qin et al. 2014) (Fig. 2 and Tables S5, S6). The discrepancy between groups I and VII (strain Mal64T) is also reflected by the different positioning of strain Mal64T in the phylogenetic trees (Fig. 1). However, the distinct clustering of the two groups in both the MLSA- and 16S rRNA sequence-based trees does not allow merging the two groups into one single genus. Moreover, the 16S rRNA gene sequence similarity between strain Mal64T and group I strains is below the 94.5% threshold for delineation of prokaryotic genera as suggested by Yarza et al. (2014).
Analysis of phylogenetic markers for delineation of genera and species. Species suspected to belong to the same species (groups I to VIII) have the same colour: (I) green, (II) red, (III) blue, (IV) yellow, (V) hot pink, (VI) light green and (VII) purple. Values determined for average nucleotide identity (ANI) can be used to differentiate between species, while amino acid identity (AAI) and percentage of conserved proteins (POCP) allow to differentiate between genera. Values obtained for rpoB and 16S rRNA gene sequence identity are available for both applications. The values are given in black within groups and in black and bold between groups, where groups might be suspected to belong together. (Minimal) identity values between well resolved groups are not given in the figure but can be found in Tables S2–S6. P. c.: Posidoniimonas corsicana; L. p.: Lacipirellula parvula; L. l.: ‘Lacipirellula limnantha’; B. g.: ‘Bythopirellula goksoyri’. (Color figure online)
Morphology and physiology
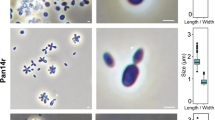
Cell morphology and cell sizes of strains Spa11T, Pla108T, Pla111T, Pla123aT, Pan181T, Pla175T, Pla144T and Mal64T were determined by observation through light microscopy and scanning electron microscopy (SEM) using exponentially growing cultures (Figs. 3, 4). All strains form pear-shaped cells, with cell sizes given in Figs. 3 and 4 and Table S7. On average, they range from 1.1 to 1.6 µm in length and 0.6 to 1.1 µm in width. All strains divide by budding, with the released buds being shaped like the mother cells. For all strains with appropriate early exponential phase data, a dimorphic life cycle could also be observed (strains Spa11T, Pla111T, Pla123aT, Pla144T). In all strains, the cells are connected by short holdfast structures that are released opposite of the budding pole. This allows most of the strains to form larger loose aggregates, with the exception of strain Mal64T (very dense aggregates) and Pla175T (only very small aggregates). For all strains but the latter, the production of fibres, that seem to foster the formation of aggregates, could be observed at the budding pole. All strains possess crateriform structures, and, at least for strains Pla111T, Pla175T and Pla144T, these structures are located at the budding pole. For most of the strains, flagella could be observed at some point of their life cycle. All strains are aerobic heterotrophs. The colonies of strains Spa11T, Pla123aT, Pan181T, Pla144T and Mal64T are non-pigmented, whereas the colonies of strains Pla108T, Pla111T, and Pla175T are coloured in hot pink to red shades. The strains were found to grow over pH ranges of 5.0 to 9.0 with the optima being between pH 6.5 and 8.5 (Table 1, Fig. S1). While most of the strains are capable of growth in a broad spectrum within this range, strain Pla111T only grew at pH 6.5–7.0. Growth of the examined strains was observed at temperatures between 10 and 39 °C with optimal growth between 24 and 30 °C (Table 1, Fig. S2). In this range, strain Pla144T had the narrowest spectrum with growth only between 20 and 30 °C (Topt of 27 °C). The growth rates at the optimal temperature in M1H NAG ASW medium were between 0.0074 and 0.041 h−1 which corresponds to doubling times between 17 and 94 h (Table 1).
Microscopy images and cell size plot of novel strains of groups I (green) and II (red). The figure shows phase contrast as well as scanning electron microscopy (SEM) micrographs. The scale bar is 1 µm. For determination of the cell sizes at least 100 representative cells were counted manually or by using a semi-automated object count tool. Whiskers of box plots represent 1.5x interquartile range. (Color figure online)
Microscopy images and cell size plot of novel strains of groups III (blue), V (hot pink), VI (light green) and VII (purple). The figure shows phase contrast as well as scanning electron microscopy (SEM) micrographs. The scale bar is 1 µm. For determination of the cell sizes at least 100 representative cells were counted manually or by using a semi-automated object count tool. Whiskers of box plots represent 1.5x interquartile range. (Color figure online)
Genome-based analysis of enzymes participating in central carbon metabolism
The genomes of all these novel strains carry genes to allow the conversion of glucose to pyruvate via the Embden-Meyerhof-Parnas pathway, but probably not via the Entner-Doudoroff pathway (Table S8). For members of groups I, II, III, V and VI it is unsure if they are capable of performing gluconeogenesis, as they might lack a pyruvate carboxylase (EC 6.4.1.1) catalysing the formation of oxaloacetate from pyruvate (Table S8). As suggested by the presence of respective genes, all strains are capable of using the pentose phosphate pathway and also the tricarboxylic acid cycle appears to be functional. Interestingly, not all strains possess the 2-oxoglutarate dehydrogenase complex (members of groups I, VI and VII as well as one member of group II). However, all strains harbour genes coding for a putative 2-oxoacid:ferredoxin oxidoreductase, that also allows the oxidation of 2-oxoglutarate to succinyl-CoA (Table S8).
Genomic features of the isolates
While the genomes of three of the strains characterised here are closed, the other genomes consist of up to 27 scaffolds (Table 2). The G+C content of the DNA varies between 58.0 and 66.5%, which brings the family Lacipirellulaceae into the upper quartile in terms of the G+C content of Planctomycetes (Table 2). The genome sizes of the strains fall between 4.33 and 6.62 Mb. Members of group I have the smallest genomes (Table 2), which are among the smallest genomes found in the class Planctomycetia. Most probably due to their smaller genomes, the number of hypothetical proteins is also relatively low throughout the family (38–44%), with a noticeable exception for L. parvula (66%). The strains have between 47 and 78 tRNAs and possess only one copy of the rRNA genes.
The determined phylogenetic groups I-VII can also be confirmed by the analysis of the pan genome of the strains (entirety of genes present in all genomes; Fig. 5). Next to genes present in all/many genomes (position 3 to 4 o’clock, Fig. 5), genes that are predominant in the separate groups support the phylogenetic assumptions: dense clusters can be found for all groups that contain more than one genome (II: red, 7 o’clock; IV: yellow, 6 o’clock;, VI: light green, 5 o’clock). At this stage, the only exception is again strain Mal64T (VII, purple) that seems to cluster with members of group I (dark green) despite their distant relationship suggested by lower 16S rRNA gene sequence similarity.
Pan genome of all strains within the family Lacipirellulaceae. Each open circle represents the pan genome of all strains, but is coloured darker when the gene is present in the respective genome. The colour represents the group affiliation of the genomes: (I) dark green, (II) red, (III) blue, (IV) yellow, (V) hot pink, (VI) light green and (VII) purple. The trees reflect the relatedness of the strains based on the absence/presence of genes. (Color figure online)
Based on the phylogenetic analysis, the absence and presence of gene clusters as well as morphological and physiological data, we propose that each group (I to VII) corresponds to a separate genus within the family Lacipirellulaceae. On the same basis, we suggest that each member of each group belongs to a separate species. Therefore, we propose the following genus and species names: (I) Botrimarina mediterranea gen. nov., sp. nov. with type strain Spa11T, Botrimarina colliarenosi sp. nov. with type strain Pla108T, Botrimarina hoheduenensis sp. nov. with type strain Pla111T; (II) Posidoniimonas polymericola sp. nov. with type strain Pla123aT; (III) Aeoliella mucimassa gen. nov., sp. nov. with type strain Pan181T; (V) Pirellulimonas nuda gen. nov., sp. nov. with type strain Pla175T; (VI) Bythopirellula polymerisocia sp. nov. with type strain Pla144T and (VII) Pseudobythopirellula maris gen. nov., sp. nov. with type strain Mal64T.
Emended description of the family Lacipirellulaceae (Dedysh et al. 2020)
The description is as given by Dedysh et al. 2020, with the following modification: The DNA G+C content of members of the genus ranges between 52 and 67%.
Description of the genus Botrimarina gen. nov.
Botrimarina (Bo.tri.ma.ri’na. L. fem. n. botrus a cluster of grapes; L. fem. adj. marina marine, of the sea; N.L. fem. n. Botrimarina a cluster of marine grapes, referring to the origin and the characteristics of the type strain of the type species).
Cells are pear-shaped and form aggregates. The DNA G+C content of the genomes of members of the genus ranges between 63 and 65%. The distinctiveness of the genus is confirmed by phylogenetic analysis. The type species is Botrimarina mediterranea. The genus belongs to the family Lacipirellulaceae, order Pirellulales, class Planctomycetia, phylum Planctomycetes.
Description of Botrimarina mediterranea sp. nov.
Botrimarina mediterranea (me.di.ter.ra’ne.a. L. fem. adj. mediterranea Mediterranean; corresponding to the origin of the strain from the Mediterranean Sea).
Cells are pear-shaped (size: 1.2 ± 0.2 × 0.6 ± 0.1 µm), form loose aggregates, divide by budding and feature a dimorphic life cycle. Cells have flagella, crateriform structures and fibres covering only one pole. On the opposite side to the latter, a holdfast structure is located. Cells are mesophilic and neutrophilic chemoorganoheterotrophs, growing over ranges of 10–36 °C (optimum 30 °C) and pH 5.0–9.5 (optimum 6.5). Colonies are white. The genome size of the type strain is 5.87 Mb with a G+C content of 64.1%.
The type strain is Spa11T (= DSM 100745T = LMG 31350T = CECT 9852T = VKM B-3431T), isolated from costal seawater at Costa Brava, Spain. The type strain genome (acc. no. CP036349) and 16S rRNA gene sequence (acc. no. MK554534) are available from GenBank.
Description of Botrimarina colliarenosi sp. nov.
Botrimarina colliarenosi (col.li.a.re.no.si. L. masc. n. collis a hill, heap; L. masc. adj. arenosus sandy; N.L. gen. n. colliarenosi from a sandy hill; corresponding to the origin of the type strain from Hohe Düne in Rostock, Germany).
Cells are pear-shaped (size: 1.2 ± 0.1 × 0.6 ± 0.1 µm), form aggregates and divide by budding. Cells have a holdfast structure, flagella, crateriform structures and polar fibres. Cells are mesophilic and neutrophilic chemoorganoheterotrophs, growing over a range of 10–33 °C (optimum 30 °C) and at pH 6.5–8.5 (optimum 6.5). Colonies are amaranth pink. The genome size of the type strain is 5.27 Mb with a DNA G+C content of 65.4 ± 3.8%.
The type strain is Pla108T (= DSM 103355T = LMG 29803T), isolated from wood particles placed in a small incubator, which was stored in a depth of two metres for 14 days close to the yacht harbour of Hohe Düne, a local district of Rostock, Germany. The type strain genome (acc. no. SJPR00000000) and 16S rRNA gene sequence (acc. no. MK554547) are available from GenBank.
Description of Botrimarina hoheduenensis sp. nov.
Botrimarina hoheduenensis (ho.he.due.nen’sis. N.L. fem. adj. hoheduenensis of Hohe Düne; corresponding to the origin of the type strain from Hohe Düne in Rostock, Germany).
Cells are pear-shaped (size: 1.3 ± 0.2 × 0.7 ± 0.1 µm), form loose aggregates, divide by budding and feature a dimorphic life cycle. Cells have flagella, crateriform structures and fibres, all emerging at the budding pole. A holdfast structure is located at the opposite pole. Cells are mesophilic and neutrophilic chemoorganoheterotrophs, growing over a range of 10–33 °C (optimum 27 °C) and at pH 6.5–7.0 (optimum 7.0). Colonies are carnation-pink pigmented. The genome size of the type strain is 4.33 Mb with a DNA G+C content of 63.1 ± 2.6%.
The type strain is Pla111T (= DSM 103485T = STH00945T, Jena Microbial Resource Collection JMRC), isolated from a biofilm formed in a small incubator, which in turn was stored in a depth of 2 metres for 14 days close to the yacht harbour of Hohe Düne, a local district of Rostock, Germany. The type strain genome (acc. no. SJPH00000000) and 16S rRNA gene sequence (acc. no. MK554579) are available from GenBank.
Emended description of the genus Posidoniimonas Kohn et al. (2020)
The description of the genus Posidoniimonas is as given previously (Kohn et al. 2020a) with the following modification: The G+C content of members of the genus is between 66.5 and 66.7%.
Description of Posidoniimonas polymericola sp. nov.
Posidoniimonas polymericola (po.ly.mer.i’co.la. N.L. neut. n. polymerum polymer; L. suff. cola an inhabitant, resident; N.L. fem. n. polymericola an inhabitant of polymers; corresponding to the origin of the type strain from polymeric material).
Cells are pear-shaped (size: 1.6 ± 0.3 × 1.0 ± 0.2 µm), form aggregates, divide by budding and feature a dimorphic life cycle. Cells have flagella, crateriform structures and polar fibres as well as a holdfast structure opposite to the fibre pole. Cells are mesophilic and neutrophilic chemoorganoheterotrophs, growing over a range of 10–33 °C (optimum 27 °C) and at pH 6.0–8.5 (optimum 7.5). Colonies are white. The genome size of the type strain is 6.26 Mb with a DNA G+C content of 66.5 ± 3.2%.
The type strain is Pla123aT (= DSM 103020T = LMG 29466T), isolated from wood particles placed in a small incubator, which was stored in a depth of 2 metres for 14 days below the pier of Heiligendamm (Seebrücke Heiligendamm), Germany. The type strain genome (acc. no. SJPO00000000) and 16S rRNA gene sequence (acc. no. MK554580) are available from GenBank.
Description of the genus Aeoliella gen. nov.
Aeoliella (Ae.o.li.el’la. N.L. fem. n. Aeoliella (dim. of L. fem. aeolia, the Eolian islands) a bacterium from the Eolian Islands, Italy).
Cells are pear-shaped and form aggregates. The distinctiveness of the genus is confirmed by phylogenetic analysis. The type species is Aeoliella mucimassa. The genus belongs to the family Lacipirellulaceae, order Pirellulales, class Planctomycetia, phylum Planctomycetes.
Description of Aeoliella mucimassa sp. nov
Aeoliella mucimassa (mu.ci.mas’sa. L. masc. n. mucus mucus, slime; L. fem. n. massa a lump; N.L. fem. n. mucimassa a mass of slime, corresponding to the characteristic of the cells to form very fibrous aggregates).
Cells are pear-shaped (size: 1.4 ± 0.3 × 0.7 ± 0.1 µm), form dense aggregates and divide by budding. Cells have crateriform structures and polar fibres. Cells are mesophilic and neutrophilic chemoorganoheterotrophs, growing over a range of 10–39 °C (optimum 28 °C) and at pH 5.5–9.0 (optimum 7.5). Colonies are white. The genome size of the type strain is 6.61 Mb with a DNA G+C content of 58.0%.
The type strain is Pan181T (= DSM 29370T = LMG 31346T = CECT 9840T = VKM B-3426T), isolated from the shallow sea hydrothermal vent system close to Panarea Island, Italy. The type strain genome (acc. no. CP036278) and 16S rRNA gene sequence (acc. no. MK559982) are available from GenBank.
Description of the genus Pirellulimonas gen. nov.
Pirellulimonas (Pi.rel.lu.li.mo’nas. N.L. fem. n. Pirellula name of a bacterial genus; L. fem. n. monas a unit, monad; N.L. fem. n. Pirellulimonas a monad of the order Pirellulales).
Cells are pear-shaped. The distinctiveness of the genus is confirmed by phylogenetic analysis. The type species is Pirellulimonas nuda. The genus belongs to the family Lacipirellulaceae, order Pirellulales, class Planctomycetia, phylum Planctomycetes.
Description of Pirellulimonas nuda sp. nov.
Pirellulimonas nuda (nu’da. L. fem. adj. nuda nude, bare; corresponding to the absence of matrix and fibers).
Cells are pear-shaped (size: 1.4 ± 0.2 × 0.8 ± 0.1 µm), form very small aggregates and divide by budding. Cells have crateriform structures located mainly at one pole. Cells are mesophilic and neutrophilic chemoorganoheterotrophs, growing over a range of 10–33 °C (optimum 24 °C) and at pH 5.5–8.5 (optimum 6.5). Colonies are ruby. The genome size of the type strain is 6.62 Mb with a DNA G+C content of 66.5%.
The type strain is Pla175T (= DSM 109594T = CECT 9871T = VKM B-3448T), isolated from wood particles placed in a small incubator, which was stored in a depth of 2 metres for 14 days in the Unterwarnow, an estuary of the river Warnow close to Rostock, Germany. The sampling location was close to a discharge of a wastewater treatment plant. The type strain genome (acc. no. CP036291) and 16S rRNA gene sequence (acc. no. MK559987) are available from GenBank.
Emended description of the genus Bythopirellula (Storesund and Øvreås 2013)
The description is as given by Storesund and Øvreås 2013, with the following modifications: Oval cells occurring assembled in consortia or as single cells. Reproducing by budding, daughter cells are motile; mother cells are non-motile and attached to substrate or surface. Produce exopolysaccharide-like structures for attachment. Member of the family Lacipirellulaceae, order Pirellulales, class Planctomycetia, phylum Planctomycetes.
Description of Bythopirellula polymerisocia sp. nov.
Bythopirellula polymerisocia (po.ly.me.ri.so’ci.a. N.L. neut. n. polymerum polymer; L. fem. adj. socia allied, united; N.L. fem. adj. polymerisocia; corresponding to the characteristic of the cells to attach to polymeric material).
Cells are pear-shaped (size: 1.1 ± 0.1 × 0.7 ± 0.1 µm), form fibrous aggregates, divide by budding and feature a dimorphic life cycle. Cells have flagella as well as polar crateriform structures and fibres. Cells are mesophilic and neutrophilic chemoorganoheterotrophs, growing over a range of 20–30 °C (optimum 27 °C) and at pH 6.0–9.5 (optimum 8.5). Colonies are white. The genome size of the type strain is 6.14 Mb with a G+C content of 52.9 ± 2.2%.
The type strain is Pla144T (= DSM 104841T = VKM B-3442T), isolated from plastic waste (polyethylene) found on the banks of the river Warnow close to Rostock, Germany. The type strain genome (acc. no. SJPS00000000) and 16S rRNA gene sequence (acc. no. MK554548) are available from GenBank.
Description of the genus Pseudobythopirellula gen. nov.
Pseudobythopirellula (Pseu.do.by.tho.pi.rel’lu.la. Gr. adj. pseudes false, mendacious; N.L. fem. n. Bythopirellula a proposed name of a bacterial genus; N.L. fem. n. Pseudobythopirellula, false Bythopirellula).
Cells are pear-shaped and form aggregates. The distinctiveness of the genus is confirmed by phylogenetic analysis. The type species is Pseudobythopirellula maris. The genus belongs to the family Lacipirellulaceae, order Pirellulales, class Planctomycetia, phylum Planctomycetes.
Description of Pseudobythopirellula maris sp. nov.
Pseudobythopirellula maris (ma’ris. L. gen. n. maris of the sea; corresponding to the origin of the type strain from the sea).
Cells are pear-shaped (size: 1.3 ± 0.2 × 0.6 ± 0.1 µm), form very dense aggregates and divide by budding. Cells have crateriform structures and form much fibrous material. Cells are mesophilic and neutrophilic chemoorganoheterotrophs, growing over a range of 15–36 °C (optimum 27 °C) and at pH 5.5–9.0 (optimum 7.0). Colonies are white. The genome size of the type strain is 5.07 Mb with a DNA G+C content of 66.2 ± 0.9%.
The type strain is Mal64T (= DSM 100832T = LMG 29020T, synonym Malle64), isolated from phytoplankton collected at a public beach on Mallorca Island, Spain. The type strain genome (acc. no. SJPQ00000000) and 16S rRNA gene sequence (acc. no. MK554544) are available from GenBank.
References
Acehan D, Santarella-Mellwig R, Devos DP (2013) A bacterial tubulovesicular network. J Cell Sci 127:277–280
Bengtsson MM, Øvreås L (2010) Planctomycetes dominate biofilms on surfaces of the kelp Laminaria hyperborea. BMC Microbiol 10:261
Bengtsson MM, Sjøtun K, Lanzén A, Øvreås L (2012) Bacterial diversity in relation to secondary production and succession on surfaces of the kelp Laminaria hyperborea. ISME J 6:2188–2198
Boedeker C, Schuler M, Reintjes G, Jeske O, van Teeseling MC, Jogler M, Rast P, Borchert D, Devos DP, Kucklick M, Schaffer M, Kolter R, van Niftrik L, Engelmann S, Amann R, Rohde M, Engelhardt H, Jogler C (2017) Determining the bacterial cell biology of Planctomycetes. Nat Commun 8:14853
Boersma AS, Kallscheuer N, Wiegand S, Rast P, Peeters SH, Mesman RJ, Heuer A, Boedeker C, Jetten MSM, Rohde M, Jogler M, Jogler C (2019) Alienimonas californiensis gen. nov. sp. nov., a novel Planctomycete isolated from the kelp forest in Monterey Bay. Antonie Van Leeuwenhoek. https://doi.org/10.1007/s10482-019-01367-4
Bondoso J, Harder J, Lage OM (2013) rpoB gene as a novel molecular marker to infer phylogeny in Planctomycetales. Antonie Van Leeuwenhoek 104:477–488
Bondoso J, Balague V, Gasol JM, Lage OM (2014) Community composition of the Planctomycetes associated with different macroalgae. FEMS Microbiol Ecol 88:445–456
Bondoso J, Albuquerque L, Nobre MF, Lobo-da-Cunha A, da Costa MS, Lage OM (2015) Roseimaritima ulvae gen. nov., sp. nov. and Rubripirellula obstinata gen. nov., sp. nov. two novel planctomycetes isolated from the epiphytic community of macroalgae. Syst Appl Microbiol 38:8–15
Bondoso J, Godoy-Vitorino F, Balague V, Gasol JM, Harder J, Lage OM (2017) Epiphytic Planctomycetes communities associated with three main groups of macroalgae. FEMS Microbiol Ecol 93:fiw255
Castresana J (2000) Selection of conserved blocks from multiple alignments for their use in phylogenetic analysis. Mol Biol Evol 17:540–552
Dedysh SN, Kulichevskaya IS, Beletsky AV, Ivanova AA, Rijpstra WIC, Damste JSS, Mardanov AV, Ravin NV (2020) Lacipirellula parvula gen. nov., sp. nov., representing a lineage of planctomycetes widespread in low-oxygen habitats, description of the family Lacipirellulaceae fam. nov. and proposal of the orders Pirellulales ord. nov., Gemmatales ord. nov. and Isosphaerales ord. nov. Syst Appl Microbiol 43:126050
Delmont TO, Eren AM (2018) Linking pangenomes and metagenomes: the Prochlorococcus metapangenome. PeerJ 6:e4320
Edgar RC (2004) MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res 32:1792–1797
Eren AM, Esen OC, Quince C, Vineis JH, Morrison HG, Sogin ML, Delmont TO (2015) Anvi’o: an advanced analysis and visualization platform for ‘omics data. PeerJ 3:e1319
Gade D, Stuhrmann T, Reinhardt R, Rabus R (2005) Growth phase dependent regulation of protein composition in Rhodopirellula baltica. Environ Microbiol 7:1074–1084
Graca AP, Calisto R, Lage OM (2016) Planctomycetes as novel source of bioactive molecules. Front Microbiol 7:1241
Jeske O, Jogler M, Petersen J, Sikorski J, Jogler C (2013) From genome mining to phenotypic microarrays: Planctomycetes as source for novel bioactive molecules. Antonie Van Leeuwenhoek 104:551–567
Jeske O, Schüler M, Schumann P, Schneider A, Boedeker C, Jogler M, Bollschweiler D, Rohde M, Mayer C, Engelhardt H, Spring S, Jogler C (2015) Planctomycetes do possess a peptidoglycan cell wall. Nat Commun 6:7116
Jeske O, Surup F, Ketteniß M, Rast P, Förster B, Jogler M, Wink J, Jogler C (2016) Developing techniques for the utilization of Planctomycetes as producers of bioactive molecules. Front Microbiol 7:1242
Jogler C, Waldmann J, Huang X, Jogler M, Glöckner FO, Mascher T, Kolter R (2012) Identification of proteins likely to be involved in morphogenesis, cell division, and signal transduction in Planctomycetes by comparative genomics. J Bacteriol 194:6419–6430
Kallscheuer N, Jogler M, Wiegand S, Peeters SH, Heuer A, Boedeker C, Jetten MSM, Rohde M, Jogler C (2019a) Rubinisphaera italica sp. nov. isolated from a hydrothermal area in the Tyrrhenian Sea close to the volcanic island Panarea. Antonie Van Leeuwenhoek. https://doi.org/10.1007/s10482-019-01329-w
Kallscheuer N, Jogler M, Wiegand S, Peeters SH, Heuer A, Boedeker C, Jetten MSM, Rohde M, Jogler C (2019b) Three novel Rubripirellula species isolated from plastic particles submerged in the Baltic Sea and the estuary of the river Warnow in northern Germany. Antonie Van Leeuwenhoek. https://doi.org/10.1007/s10482-019-01368-3
Kallscheuer N, Wiegand S, Jogler M, Boedeker C, Peeters SH, Rast P, Heuer A, Jetten MSM, Rohde M, Jogler C (2019c) Rhodopirellula heiligendammensis sp. nov., Rhodopirellula pilleata sp. nov., and Rhodopirellula solitaria sp. nov. isolated from natural or artificial marine surfaces in Northern Germany and California, USA, and emended description of the genus Rhodopirellula. Antonie Van Leeuwenhoek. https://doi.org/10.1007/s10482-019-01366-5
Kallscheuer N, Wiegand S, Peeters SH, Jogler M, Boedeker C, Heuer A, Rast P, Jetten MSM, Rohde M, Jogler C (2019d) Description of three bacterial strains belonging to the new genus Novipirellula gen. nov., reclassificiation of Rhodopirellula rosea and Rhodopirellula caenicola and readjustment of the genus threshold of the phylogenetic marker rpoB for Planctomycetaceae. Antonie Van Leeuwenhoek. https://doi.org/10.1007/s10482-019-01374-5
Kallscheuer N, Jeske O, Sandargo B, Boedeker C, Wiegand S, Bartling P, Jogler M, Rohde M, Petersen J, Medema MH, Surup F, Jogler C (2020a) The planctomycete Stieleria maiorica Mal15T employs stieleriacines to alter the species composition in marine biofilms. Commun Biol 3:303
Kallscheuer N, Wiegand S, Boedeker C, Peeters SH, Jogler M, Heuer A, Jetten MS, Rohde M, Jogler C (2020b) Caulifigura coniformis gen. nov., sp. nov., a novel member of the family Planctomycetaceae isolated from a red biofilm sampled in a hydrothermal area. Antonie Van Leeuwenhoek. https://doi.org/10.1007/s10482-020-01439-w
Kallscheuer N, Wiegand S, Boedeker C, Peeters SH, Jogler M, Rast P, Heuer A, Jetten MSM, Rohde M, Jogler C (2020c) Aureliella helgolandensis gen. nov., sp. nov., a novel Planctomycete isolated from a jellyfish at the shore of the island Helgoland. Antonie Van Leeuwenhoek. https://doi.org/10.1007/s10482-020-01403-8
Kallscheuer N, Wiegand S, Heuer A, Rensink S, Boersma AS, Jogler M, Boedeker C, Peeters SH, Rast P, Jetten MSM, Rohde M, Jogler C (2020d) Blastopirellula retiformator sp. nov. isolated from the shallow-sea hydrothermal vent system close to Panarea Island. Antonie Van Leeuwenhoek. https://doi.org/10.1007/s10482-019-01377-2
Kallscheuer N, Rast P, Jogler M, Wiegand S, Kohn T, Boedeker C, Jeske O, Heuer A, Quast C, Glöckner FO, Rohde M, Jogler C (in revision) Analysis of bacterial communities in a municipal duck pond during a phytoplankton bloom and isolation of three novel strains of the phylum Planctomycetes. Environ Microbiol
Kim M, Oh HS, Park SC, Chun J (2014) Towards a taxonomic coherence between average nucleotide identity and 16S rRNA gene sequence similarity for species demarcation of prokaryotes. Int J Syst Evol Microbiol 64:346–351
Kohn T, Rast P, Wiegand S, Jetten M, Kallscheuer N, Jeske O, Boedeker C, Rohde M, Jogler M, Jogler C (2020a) The microbiome of Posidonia oceanica seagrass leaves can be dominated by Planctomycetes. Front Microbiol 11:1458
Kohn T, Wiegand S, Boedeker C, Rast P, Heuer A, Jetten MSM, Schuler M, Becker S, Rohde C, Muller RW, Brummer F, Rohde M, Engelhardt H, Jogler M, Jogler C (2020b) Planctopirus ephydatiae, a novel Planctomycete isolated from a freshwater sponge. Syst Appl Microbiol 43:126022
Kumar D, Gaurav K, Pk S, Uppada J, Ch VR (2020) Gimesia chilikensis sp. nov., a haloalkali-tolerant planctomycete isolated from Chilika lagoon and emended description of the genus Gimesia. Int J Syst Evol Microbiol 70:3647–3655
Lachnit T, Fischer M, Kunzel S, Baines JF, Harder T (2013) Compounds associated with algal surfaces mediate epiphytic colonization of the marine macroalga Fucus vesiculosus. FEMS Microbiol Ecol 84:411–420
Lage OM, Bondoso J (2014) Planctomycetes and macroalgae, a striking association. Front Microbiol 5:267
Lechner M, Findeiss S, Steiner L, Marz M, Stadler PF, Prohaska SJ (2011) Proteinortho: detection of (co-)orthologs in large-scale analysis. BMC Bioinform 12:124
Lee I, Ouk Kim Y, Park SC, Chun J (2016) OrthoANI: an improved algorithm and software for calculating average nucleotide identity. Int J Syst Evol Microbiol 66:1100–1103
Luo C, Rodriguez RL, Konstantinidis KT (2014) MyTaxa: an advanced taxonomic classifier for genomic and metagenomic sequences. Nucleic Acids Res 42:e73
Mitchell AL, Attwood TK, Babbitt PC, Blum M, Bork P, Bridge A, Brown SD, Chang HY, El-Gebali S, Fraser MI, Gough J, Haft DR, Huang H, Letunic I, Lopez R, Luciani A, Madeira F, Marchler-Bauer A, Mi H, Natale DA, Necci M, Nuka G, Orengo C, Pandurangan AP, Paysan-Lafosse T, Pesseat S, Potter SC, Qureshi MA, Rawlings ND, Redaschi N, Richardson LJ, Rivoire C, Salazar GA, Sangrador-Vegas A, Sigrist CJA, Sillitoe I, Sutton GG, Thanki N, Thomas PD, Tosatto SCE, Yong SY, Finn RD (2019) InterPro in 2019: improving coverage, classification and access to protein sequence annotations. Nucleic Acids Res 47:D351–D360
Peeters S, Wiegand S, Kallscheuer N, Jogler M, Heuer A, Jetten M, Rast P, Boedeker C, Rohde M, Jogler C (2019) Three marine strains constitute the novel genus and species Crateriforma conspicua in the phylum Planctomycetes. Antonie Van Leeuwenhoek. https://doi.org/10.1007/s10482-019-01375-4
Peeters SH, Wiegand S, Kallscheuer N, Jogler M, Heuer A, Jetten MS, Boedeker C, Rohde M, Jogler C (2020) Description of Polystyrenella longa gen. nov., sp. nov., isolated from polystyrene particles incubated in the Baltic Sea. https://doi.org/10.1007/s10482-020-01406-5
Pilhofer M, Rappl K, Eckl C, Bauer AP, Ludwig W, Schleifer KH, Petroni G (2008) Characterization and evolution of cell division and cell wall synthesis genes in the bacterial phyla Verrucomicrobia, Lentisphaerae, Chlamydiae, and Planctomycetes and phylogenetic comparison with rRNA genes. J Bacteriol 190:3192–3202
Price MN, Dehal PS, Arkin AP (2010) FastTree 2—approximately maximum-likelihood trees for large alignments. PLoS ONE 5:e9490
Pruesse E, Peplies J, Glöckner FO (2012) SINA: accurate high-throughput multiple sequence alignment of ribosomal RNA genes. Bioinformatics 28:1823–1829
Qin Q-L, Xie B-B, Zhang X-Y, Chen X-L, Zhou B-C, Zhou J, Oren A, Zhang Y-Z (2014) A proposed genus boundary for the prokaryotes based on genomic insights. J Bacteriol 196:2210–2215
Rast P, Glöckner I, Boedeker C, Jeske O, Wiegand S, Reinhardt R, Schumann P, Rohde M, Spring S, Glöckner FO, Jogler C, Jogler M (2017) Three novel species with peptidoglycan cell walls form the new genus Lacunisphaera gen. nov. in the family Opitutaceae of the verrucomicrobial subdivision 4. Front Microbiol 8:202
Rensink S, Wiegand S, Kallscheuer N, Rast P, Peeters SH, Heuer A, Boedeker C, Jetten MSM, Rohde M, Jogler M, Jogler C (2020) Description of the novel planctomycetal genus Bremerella, containing Bremerella volcania sp. nov., isolated from an active volcanic site, and reclassification of Blastopirellula cremea as Bremerella cremea comb. nov. Antonie Van Leeuwenhoek. https://doi.org/10.1007/s10482-019-01378-1
Rivas-Marin E, Wiegand S, Kallscheuer N, Jogler M, Peeters SH, Heuer A, Jetten MS, Boedeker C, Rohde M, Devos DP, Jogler C (2020a) Maioricimonas rarisocia gen. nov., sp. nov., a novel planctomycete isolated from marine sediments close to Mallorca Island. Antonie Van Leeuwenhoek. https://doi.org/10.1007/s10482-020-01436-z
Rivas-Marin E, Wiegand S, Kallscheuer N, Jogler M, Peeters SH, Heuer A, Jetten MS, Boedeker C, Rohde M, Devos DP, Jogler C (2020b) Thalassoglobus polymorphus sp. nov., a novel Planctomycete isolated close to a public beach of Mallorca Island. Antonie Van Leeuwenhoek. https://doi.org/10.1007/s10482-020-01437-y
Rodriguez-R LM, Konstantinidis KT (2016) The enveomics collection: a toolbox for specialized analyses of microbial genomes and metagenomes. PeerJ 4:e:1900v1
Schubert T, Kallscheuer N, Wiegand S, Boedeker C, Peeters SH, Jogler M, Heuer A, Jetten MSM, Rohde M, Jogler C (2020) Calycomorphotria hydatis gen. nov., sp. nov., a novel species in the family Planctomycetaceae with conspicuous subcellular structures. Antonie Van Leeuwenhoek. https://doi.org/10.1007/s10482-020-01419-0
Sievers F, Wilm A, Dineen D, Gibson TJ, Karplus K, Li W, Lopez R, McWilliam H, Remmert M, Söding J (2011) Fast, scalable generation of high-quality protein multiple sequence alignments using Clustal Omega. Mol Syst Biol 7:539
Slobodkina GB, Kovaleva OL, Miroshnichenko ML, Slobodkin AI, Kolganova TV, Novikov AA, van Heerden E, Bonch-Osmolovskaya EA (2015) Thermogutta terrifontis gen. nov., sp. nov. and Thermogutta hypogea sp. nov., thermophilic anaerobic representatives of the phylum Planctomycetes. Int J Syst Evol Microbiol 65:760–765
Slobodkina GB, Panteleeva AN, Beskorovaynaya DA, Bonch-Osmolovskaya EA, Slobodkin AI (2016) Thermostilla marina gen. nov., sp. nov., a thermophilic, facultatively anaerobic planctomycete isolated from a shallow submarine hydrothermal vent. Int J Syst Evol Microbiol 66:633–638
Stamatakis A (2014) RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics 30:1312–1313
Storesund JE, Øvreås L (2013) Diversity of Planctomycetes in iron-hydroxide deposits from the Arctic Mid Ocean Ridge (AMOR) and description of Bythopirellula goksoyri gen. nov., sp. nov., a novel Planctomycete from deep sea iron-hydroxide deposits. Antonie Van Leeuwenhoek 104:569–584
UniProt C (2019) UniProt: a worldwide hub of protein knowledge. Nucleic Acids Res 47:D506–D515
van Teeseling MC, Mesman RJ, Kuru E, Espaillat A, Cava F, Brun YV, VanNieuwenhze MS, Kartal B, van Niftrik L (2015) Anammox Planctomycetes have a peptidoglycan cell wall. Nat Commun 6:6878
Vollmers J, Frentrup M, Rast P, Jogler C, Kaster AK (2017) Untangling genomes of novel planctomycetal and verrucomicrobial species from monterey bay kelp forest metagenomes by refined binning. Front Microbiol 8:472
Wagner M, Horn M (2006) The Planctomycetes, Verrucomicrobia, Chlamydiae and sister phyla comprise a superphylum with biotechnological and medical relevance. Curr Opin Biotechnol 17:241–249
Waqqas M, Salbreiter M, Kallscheuer N, Jogler M, Wiegand S, Heuer A, Rast P, Peeters SH, Boedeker C, Jetten MSM, Rohde M, Jogler C (2020) Rosistilla oblonga gen. nov., sp. nov. and Rosistilla carotiformis sp. nov., isolated from biotic or abiotic surfaces in Northern Germany, Mallorca, Spain and California, USA. Antonie Van Leeuwenhoek. https://doi.org/10.1007/s10482-020-01441-2
Wiegand S, Jogler M, Jogler C (2018) On the maverick Planctomycetes. FEMS Microbiol Rev 42:739–760
Wiegand S, Jogler M, Boedeker C, Pinto D, Vollmers J, Rivas-Marín E, Kohn T, Peeters SH, Heuer A, Rast P, Oberbeckmann S, Bunk B, Jeske O, Meyerdierks A, Storesund JE, Kallscheuer N, Lücker S, Lage OM, Pohl T, Merkel BJ, Hornburger P, Müller R-W, Brümmer F, Labrenz M, Spormann AM, Op den Camp H, Overmann J, Amann R, Jetten MSM, Mascher T, Medema MH, Devos DP, Kaster A-K, Øvreås L, Rohde M, Galperin MY, Jogler C (2020) Cultivation and functional characterization of 79 Planctomycetes uncovers their unique biology. Nat Microbiol 5:126–140
Will SE, Henke P, Boedeker C, Huang S, Brinkmann H, Rohde M, Jarek M, Friedl T, Seufert S, Schumacher M (2018) Day and night: metabolic profiles and evolutionary relationships of six axenic non-marine cyanobacteria. Genome Biol Evol 11:270–294
Yarza P, Yilmaz P, Pruesse E, Glöckner FO, Ludwig W, Schleifer KH, Whitman WB, Euzeby J, Amann R, Rossello-Mora R (2014) Uniting the classification of cultured and uncultured bacteria and archaea using 16S rRNA gene sequences. Nat Rev Microbiol 12:635–645
Acknowledgements
Part of this research was funded by the Deutsche Forschungsgemeinschaft grants KA 4967/1-1 and JO 893/4-1, grant ALWOP.308 of the Nederlandse Organisatie voor Wetenschappelijk Onderzoek (NWO), SIAM (Soehngen Institute for Anaerobic Microbiology) Grant No. 024002002 and the Radboud Excellence fellowship. We thank Ina Schleicher for skillful technical assistance. Brian Tindall and Regine Fähnrich from the DSMZ as well as the staff from the culture collections in Belgium, Spain and Russia we thank for support during strain deposition. We also thank our collaborators Sonja Oberbeckmann and Matthias Labrenz (IOW Warnemünde, Germany) as well as the Scientific Diving Center of the Bergakademie Freiberg, Germany, Thomas Pohl, Peter Hornburger and all participants of the 2013 Panarea Expedition for sampling support.
Funding
Open Access funding enabled and organized by Projekt DEAL.
Author information
Authors and Affiliations
Contributions
SW wrote the first draft of the manuscript and prepared the figures, MJ and SW performed the genomic and phylogenetic analysis, AH isolated the strains and performed the initial cultivation and strain deposition, SHP and CB performed the light microscopic analysis, MSMJ, NK and A-KK contributed to data analysis, text preparation and revised the manuscript, MR performed the electron microscopic analysis, CJ took the samples, supervised AH and the study. All authors read and approved the final version of the manuscript.
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Ethical approval
This article does not contain any studies with animals performed by any of the authors.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Wiegand, S., Jogler, M., Boedeker, C. et al. Updates to the recently introduced family Lacipirellulaceae in the phylum Planctomycetes: isolation of strains belonging to the novel genera Aeoliella, Botrimarina, Pirellulimonas and Pseudobythopirellula and the novel species Bythopirellula polymerisocia and Posidoniimonas corsicana. Antonie van Leeuwenhoek 113, 1979–1997 (2020). https://doi.org/10.1007/s10482-020-01486-3
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10482-020-01486-3