Abstract
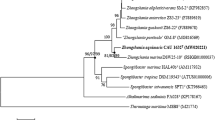
Strain K001 was isolated from a cyanobacterial culture derived from Abrolhos, a reef bank microbial mat (South Atlantic Ocean—Brazil). Cells of K001 are Gram stain–negative, catalase and oxidase-positive, non-motile, rod-shaped, and with or without appendages. Phylogenetic analysis based on 16S rRNA gene sequences showed that strain K001 belongs to the genus Muricauda. The highest strain K001 16S rRNA gene identity, ANI, and dDDH, respectively, are with M. aquimarina (98.90%, 79.23, 21.60%), M. ruestringensis (98.20%, 80.82, 23.40%), and M. lutimaris (97.86%, 79.23, 22.70%). The strain grows at 15–37 °C and between 0.5 and 10% NaCl. The major fatty acids of strain K001 are iso-C15:0, iso-C15:1 G, iso-C17:0 3-OH, and summed feature 3 (C16:1 ω6c and/or C16:1 ω7c). The polar lipids are represented by phosphatidylethanolamine, three unidentified aminolipids, and three unidentified polar lipids. The major respiratory quinone is MK-6. The G+C content of the DNA of strain K001 is 41.62 mol%. Based on polyphasic analysis of strain K001, it was identified as a novel representative of the genus Muricauda and was named Muricauda brasiliensis sp. nov. The type strain is K001 (=CBMAI 2315T = CBAS 752T).



Similar content being viewed by others
Abbreviations
- MA:
-
marine agar DIFCO 2216
- MB:
-
marine broth DIFCO 2216
- MALDI-TOF:
-
matrix-assisted laser desorption/ionization
- MS:
-
mass spectrogram
- dDDH:
-
digital DNA–DNA hybridization
- ANI:
-
average nucleotide identity
- TYGS:
-
Type Strain Genome Server
- MIDI:
-
Sherlock Microbial Identification System
References
Bruns A, Rohde M, Berthe-Corti L (2001) Muricauda ruestringensis gen. nov., sp. nov., a facultatively anaerobic, appendaged bacterium from German North Sea intertidal sediment. Int J Syst Evol Microbiol 51:1997–2006. https://doi.org/10.1099/00207713-51-6-1997
Wu YH, Yu PS, Zhou YD, Xu L, Wang CS, Wu M, Oren A, Xu XW (2013) Muricauda antarctica sp. nov., a marine member of the Flavobacteriaceae isolated from Antarctic seawater. Int J Syst Evol Microbiol 63:3451–3456. https://doi.org/10.1099/ijs.0.048355-0
Su Y, Yang X, Wang Y, Liu Y, Ren Q, Zhang XH (2017) Muricauda marina sp. Nov., isolated from marine snow of yellow sea. Int J Syst Evol Microbiol 67:2446–2451. https://doi.org/10.1099/ijsem.0.001992
Dang Y, Sun Y, Sun L, Yuan X, Li Y, Qin Q et al (2019) Muricauda nanhaiensis sp. nov., isolated from seawater of the South China Sea. Int J Syst Evol Microbiol 69:2089–2094. https://doi.org/10.1099/ijsem.0.003437
Hwang CY, Kim MH, Bae GD, Zhang GI, Kim YH, Cho BC (2009) Muricauda olearia sp. nov., isolated from crude-oil-contaminated seawater, and emended description of the genus Muricauda. Int J Syst Evol Microbiol 59:1856–1861. https://doi.org/10.1099/ijs.0.007708-0
Zhang Z, Gao X, Qiao Y, Wang Y, Zhang XH (2015) Muricauda pacifica sp. Nov., isolated from seawater of the South Pacific Gyre. Int J Syst Evol Microbiol 65:4087–4092. https://doi.org/10.1099/ijsem.0.000542
Kim JM, Jin HM, Jeon CO (2013) Muricauda taeanensis sp. nov., isolated from a marine tidal flat. Int J Syst Evol Microbiol 63:2672–2677. https://doi.org/10.1099/ijs.0.047647-0
Yang C, Li Y, Guo Q, Lai Q, Wei J, Zheng T, Tian Y (2013) Muricauda zhangzhouensis sp. nov., isolated from mangrove sediment. Int J Syst Evol Microbiol 63:2320–2325. https://doi.org/10.1099/ijs.0.040881-0
Li G, Lai Q, Yan P, Shao Z (2019) Roseovarius amoyensis sp. nov. and Muricauda amoyensis sp. nov., isolated from the Xiamen coast. Int J Syst Evol Microbiol 69:3093–3101. https://doi.org/10.1099/ijsem.0.003595
Liu L, Yu M, Zhou S, Fu T, Sun W, Wang L et al (2020) Muricauda alvinocaridis sp. nov., isolated from shrimp gill from the Okinawa Trough. Int J Syst Evol Microbiol 70:1666–1671. https://doi.org/10.1099/ijsem.0.003953
Lee SY, Park S, Oh TK, Yoon JH (2012) Muricauda beolgyonensis sp. nov., isolated from a tidal flat. Int J Syst Evol Microbiol 62:1134–1139. https://doi.org/10.1099/ijs.0.032581-0
Yoon JH, Lee MH, Oh TK, Park YH (2005) Muricauda flavescens sp. nov. and Muricauda aquimarina sp. nov., isolated from a salt lake near Hwajinpo Beach of the East Sea in Korea, and emended description of the genus Muricauda. Int J Syst Evol Microbiol 55:1015–1019. https://doi.org/10.1099/ijs.0.03051-0
Park JS (2019) Muricauda hymeniacidonis sp. Nov., isolated from sponge of Hymeniacidon sinapium. Int J Syst Evol Microbiol 69:3800–3805. https://doi.org/10.1099/ijsem.0.003683
Zhang X, Liu X, Lai Q, Du Y, Sun F, Shao Z (2018) Muricauda indica sp. nov., isolated from deep sea water. Int J Syst Evol Microbiol 68:881–885. https://doi.org/10.1099/IJSEM.0.002602
Liu SQ, Sun QL, Sun YY, Yu C, Sun L (2018) Muricauda iocasae sp. Nov., isolated from deep sea sediment of the South China Sea. Int J Syst Evol Microbiol 68:2538–2544. https://doi.org/10.1099/ijsem.0.002870
Arun AB, Chen WM, Lai WA, Chao JH, Rekha PD, Shen FT, Singh S, Young CC (2009) Muricauda lutaonensis sp. nov., a moderate thermophile isolated from a coastal hot spring. Int J Syst Evol Microbiol 59:2738–2742. https://doi.org/10.1099/ijs.0.007930-0
Wang Y, Yang X, Liu J, Wu Y, Zhang XH (2017) Muricauda lutea sp. nov., isolated from seawater. Int J Syst Evol Microbiol 67:1064–1069. https://doi.org/10.1099/ijsem.0.001792
Yoon JH, Kang SJ, Jung YT, Oh TK (2008) Muricauda lutimaris sp. nov., isolated from a tidal flat of the Yellow Sea. Int J Syst Evol Microbiol 58:1603–1607. https://doi.org/10.1099/ijs.0.65659-0
Weisburg WG, Barns SM, Pelletier DA, Lane DJ (1991) 16S ribosomal DNA amplification for phylogenetic study. J Bacteriol 173:697–703. https://doi.org/10.1128/JB.173.2.697-703.1991
Yoon SH, Ha SM, Kwon S, Lim J, Kim Y, Seo H, Chun J (2017) Introducing EzBioCloud: a taxonomically united database of 16S rRNA gene sequences and whole-genome assemblies. Int J Syst Evol Microbiol 67:1613–1617. https://doi.org/10.1099/ijsem.0.001755
Kumar S, Stecher G, Tamura K (2016) MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol Biol Evol 33:1870–1874. https://doi.org/10.1093/molbev/msw054
Thompson JD, Gibson TJ, Plewniak F, Jeanmougin F, Higgins DG (1997) The CLUSTAL X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res 25:4876–4882. https://doi.org/10.1093/nar/25.24.4876
Felsenstein J (1985) Confidence limits on phylogenies: an approach using the bootstrap. Evolution (N Y) 39:783–791. https://doi.org/10.1111/j.1558-5646.1985.tb00420.x
Saitou N, Nei M (1987) The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol 4:406–425. https://doi.org/10.1093/oxfordjournals.molbev.a040454
Kimura M (1980) A simple method for estimating evolutionary rates of base substitutions through comparative studies of nucleotide sequences. J Mol Evol 16:111–120. https://doi.org/10.1007/BF01731581
Vizzotto CS, Lopes FAC, Green SJ, Steindorff AS, Walter JM, Thompson FL, Krüger RH (2018) Draft genome sequence of Muricauda sp. strain K001 isolated from a marine cyanobacterial culture. Genome Announc 6:1–2. https://doi.org/10.1128/genomeA.00451-18
Chun J, Oren A, Ventosa A, Christensen H, Arahal DR, da Costa MS, Rooney AP, Yi H, Xu XW, de Meyer S, Trujillo ME (2018) Proposed minimal standards for the use of genome data for the taxonomy of prokaryotes. Int J Syst Evol Microbiol 68:461–466. https://doi.org/10.1099/ijsem.0.002516
Chun J, Rainey FA (2014) Integrating genomics into the taxonomy and systematics of the Bacteria and Archaea. Int J Syst Evol Microbiol 64:316–324. https://doi.org/10.1099/ijs.0.054171-0
Yoon SH, Ha S, Min LJ, Kwon S, Chun J (2017) A large-scale evaluation of algorithms to calculate average nucleotide identity. Antonie van Leeuwenhoek. Int J Gen Mol Microbiol 110:1281–1286. https://doi.org/10.1007/s10482-017-0844-4
Meier-Kolthoff JP, Auch AF, Klenk HP, Göker M (2013) Genome sequence-based species delimitation with confidence intervals and improved distance functions. BMC Bioinformatics 14:60 https://doi.org/10.1186/1471-2105-14-60
Parks DH, Imelfort M, Skennerton CT, Hugenholtz P, Tyson GW (2015) CheckM: assessing the quality of microbial genomes recovered from isolates, single cells, and metagenomes. Genome Res 25:1043–1055. https://doi.org/10.1101/gr.186072.114
Li H, Durbin R (2009) Fast and accurate short read alignment with burrows-wheeler transform. Bioinformatics 25:1754–1760. https://doi.org/10.1093/bioinformatics/btp324
Wu M, Eisen JA (2008) A simple, fast, and accurate method of phylogenomic inference. Genome Biol 9:R151. https://doi.org/10.1186/gb-2008-9-10-r151
Katoh K, Standley DM (2013) MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol 30:772–780. https://doi.org/10.1093/molbev/mst010
Price MN, Dehal PS, Arkin AP (2009) Fasttree: computing large minimum evolution trees with profiles instead of a distance matrix. Mol Biol Evol 26:1641–1650. https://doi.org/10.1093/molbev/msp077
Ha S-M, Kim CK, Roh J, Byun J-H, Yang S-J, Choi S-B, Chun J, Yong D (2019) Application of the whole genome-based bacterial identification system, TrueBac ID, using clinical isolates that were not identified with three matrix-assisted laser desorption/ionization time-of-flight mass spectrometry (MALDI-TOF MS) systems. Ann Lab Med 39:530–536. https://doi.org/10.3343/alm.2019.39.6.530
Meier-Kolthoff JP, Göker M (2019) TYGS is an automated high-throughput platform for state-of-the-art genome-based taxonomy. Nat Commun 10:2182. https://doi.org/10.1038/s41467-019-10210-3
Reddy, Beveridge, Breznak, Marzluf, Schmidt, Snyder, editors. Methods for general and molecular microbiology, Third Edition. 3a edition. American Society of Microbiology; 2007
Souza W (ed) (2007). Técnicas de microscopia eletrônica aplicadas às ciências biológicas. 3a edição. Sociedade Brasileira de Microscopia, Rio de Janeiro
Raney F, Oren A (2011) Methods in microbiology. In: Recent titles in the series, vol 38. Academic Press, London
Bernardet JF, Segers P, Vancanneyt M, Berthe F, Kersters K, Vandamme P (1996) Cutting a gordian knot: emended classification and description of the genus Flavobacterium, emended description of the family Flavobacteriaceae, and proposal of Flavobacterium hydatis nom. nov. (basonym, Cytophaga aquatilis Strohl and Tait 1978). Int J Syst Bacteriol 46:128–148. https://doi.org/10.1099/00207713-46-1-128
Tindall BJ (1990) Lipid composition of Halobacterium lacusprofundi. FEMS Microbiol Lett 66:199–202. https://doi.org/10.1016/0378-1097(90)90282-U
Tindall BJ (1990) A comparative study of the lipid composition of Halobacterium saccharovorum from various sources. Syst Appl Microbiol 13:128–130. https://doi.org/10.1016/S0723-2020(11)80158-X
Bligh EG, Dyer WJ (1959) A rapid method of total lipid extraction and purification. Can J Biochem Physiol 37:911–917. https://doi.org/10.1139/o59-099
Sasser M (1990) Fatty acid profiling by gas chromatography. Tech. Note 101 - MIDI, Newark
(2015) Fatty acid profiling by gas chromatography - for the Sherlock MIS. Microbial ID, Inc., Newak
Ramasamy D, Mishra AK, Lagier JC, Padhmanabhan R, Rossi M, Sentausa E, Raoult D, Fournier PE (2014) A polyphasic strategy incorporating genomic data for the taxonomic description of novel bacterial species. Int J Syst Evol Microbiol 64:384–391. https://doi.org/10.1099/ijs.0.057091-0
Vandamme P, Peeters C (2014) Time to revisit polyphasic taxonomy. Antonie van Leeuwenhoek. Int J Gen Mol Microbiol 106:57–65. https://doi.org/10.1007/s10482-014-0148-x
Agustini BC, Silva LP, Bloch C, Bonfim TMB, Da Silva GA (2014) Evaluation of MALDI-TOF mass spectrometry for identification of environmental yeasts and development of supplementary database. Appl Microbiol Biotechnol 98:5645–5654. https://doi.org/10.1007/s00253-014-5686-7
Schumann P, Maier T. MALDI-TOF mass spectrometry applied to classification and identification of bacteria. vol. 41. 1st ed. Elsevier Ltd.; 2014. https://doi.org/10.1016/bs.mim.2014.06.002
Prabhu S, Rekha PD, Arun AB (2014) Zeaxanthin biosynthesis by members of the genus Muricauda. Polish J Microbiol 63:115–119. https://doi.org/10.33073/pjm-2014-017
Prabhu S, Pd R, Young CC, Hameed A, Lin SY, Ab A (2013) Zeaxanthin production by novel marine isolates from coastal sand of India and its antioxidant properties. Appl Biochem Biotechnol 171:817–831. https://doi.org/10.1007/s12010-013-0397-6
Acknowledgments
We thank the Laboratory of Environmental Sanitation, Department of Civil and Environmental Engineering, University of Brasilia (Brazil), for the infrastructure to maintain the cultures and to perform some analyses. We thank the DNA Services Facility in Research Resources Center, University of Illinois at Chicago, Chicago (USA) for the sequencing service. We are grateful to the Microscopy and Microanalysis Laboratory of the University of Brasilia (Brazil), for their collaboration in carrying out the scanning electron microscopy analysis. We thank the Laboratory of Toxinology (Department of Physiological Sciences, Biological Sciences Institute, University of Brasilia, Brazil), the Laboratories of Graduate Program in Genomics Science and Biotechnology (Catholic University of Brasilia, Brasilia, Brazil), Laboratory of Mass Spectrometry (EMBRAPA Genetic Resources and Biotechnology, Brasilia, Brazil), and the Laboratories of the Department of Microbiology (Federal University of Viçosa, Viçosa, Brazil) for the infrastructure to perform part of the experiments of this work. We thank Dr. Manuela da Silva for the K001 microbial collections deposition efforts.
Funding
This research was supported by a grant from BioTecMar (no. 408339/2013-6) of CNPq (Ministry of Science, Technology, Innovations and Communications—Brazil), the Foundation for Research Support of the Federal District (FAP-DF), and Coordination for the Improvement of Higher Education Personnel (CAPES).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflicts of interest.
Additional information
Responsible Editor: Jorge Luiz Mello Sampaio
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Accession numbers: The GenBank/EMBL/DDBJ accession numbers of K001T strain for the 16S rRNA gene sequence is MN996941, and the draft genome sequence is QBTW00000000.
Rights and permissions
About this article
Cite this article
Vizzotto, C.S., Peixoto, J., Green, S.J. et al. Muricauda brasiliensis sp. nov., isolated from a mat-forming cyanobacterial culture. Braz J Microbiol 52, 325–333 (2021). https://doi.org/10.1007/s42770-020-00400-3
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s42770-020-00400-3