Abstract
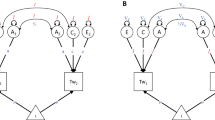
Objective: To explore and apply multimodel inference to test the relative contributions of latent genetic, environmental and direct causal factors to the covariation between two variables with data from the classical twin design by estimating model-averaged parameters. Methods: Behavior genetics is concerned with understanding the causes of variation in phenotypes and the causes of covariation between two or more phenotypes. Two variables may correlate as a result of genetic, shared environmental or unique environmental factors contributing to variation in both variables. Two variables may also correlate because one or both directly cause variation in the other. Furthermore, covariation may result from any combination of these sources, leading to 25 different identified structural equation models. OpenMx was used to fit all these models to account for covariation between two variables collected in twins. Multimodel inference and model averaging were used to summarize the key sources of covariation, and estimate the magnitude of these causes of covariance. Extensions of these models to test heterogeneity by sex are discussed. Results: We illustrate the application of multimodel inference by fitting a comprehensive set of bivariate models to twin data from the Virginia Twin Study of Psychiatric and Substance Use Disorders. Analyses of body mass index and tobacco consumption data show sufficient power to reject distinct models, and to estimate the contribution of each of the five potential sources of covariation, irrespective of selecting the best fitting model. Discrimination between models on sample size, type of variable (continuous versus binary or ordinal measures) and the effect size of sources of variance and covariance. Conclusions: We introduce multimodel inference and model averaging approaches to the behavior genetics community, in the context of testing models for the causes of covariation between traits in term of genetic, environmental and causal explanations.




Similar content being viewed by others
References
Boker S et al (2011) OpenMx: an open source extended structural equation modeling framework. Psychometrika 76(2):306–317
Burnham KP, Anderson DR (2004) Multimodel inference—understanding AIC and BIC in model selection. Sociol Methods Res 33(2):261–304
Carey G (2005) Cholesky problems. Behav Genet 35(5):653–665
Duffy DL, Martin NG (1994) Inferring the direction of causation in cross-sectional twin data: theoretical and empirical considerations. Genet Epidemiol 11(6):483–502
Heath AC et al (1993) Testing hypotheses about direction of causation using cross-sectional family data. Behav Genet 23(1):29–50
Kendler KS, Prescott CA (2006) Genes, environment, and psychopathology: understanding the causes of psychiatric and substance use disorders. Guilford Press, New York
Kirkpatrick RM et al (2015) Replication of a gene–environment interaction via multimodel inference: additive-genetic variance in adolescents’ general cognitive ability increases with family-of-origin socioeconomic status. Behav Genet 45(2):200–214
Maes HH (2005) The ACE model. In: Purcell S (ed) Encyclopedia for behavioral statistics. Wiley series in probability and statistics. Wiley, New York
Maes HH (2018) OpenMx Scripts. https://heremine.maes/squarespace.com
Maes HH et al (2004) A twin study of genetic and environmental influences on tobacco initiation, regular tobacco use and nicotine dependence. Psychol Med 34(7):1251–1261
Neale MC et al (1994) The power of the classical twin study to resolve variation in threshold traits. Behav Genet 24(3):239–258
Neale MC et al (2006a) Methodological issues in the assessment of substance use phenotypes. Addict Behav 31(6):1010–1034
Neale MC et al (2006b) Extensions to the modeling of initiation and progression: applications to substance use and abuse. Behav Genet 36(4):507–524
Neale MC et al (2006c) Multivariate genetic analysis of sex limitation and G × E interaction. Twin Res Hum Genet 9(4):481–489
Neale MC et al (2016) OpenMx 2.0: extended structural equation and statistical modeling. Psychometrika 81(2):535–549
Neale MC, Cardon LR (1992) Methodology for genetic studies of twins and families. Kluwer, Dordrecht
Neale MC, Miller MB (1997) The use of likelihood-based confidence intervals in genetic models. Behav Genet 27(2):113–120
Symonds MRE, Moussalli A (2011) A brief guide to model selection, multimodel inference and model averaging in behavioural ecology using Akaike’s information criterion. Behav Ecol Sociobiol 65(1):13–21
Verhulst B et al (2019) Type I error rates and parameter bias in multivariate behavioral genetic models. Behav Genet 49(1):99–111
Visscher PM (2006) A note on the asymptotic distribution of likelihood ratio tests to test variance components. Twin Res Hum Genet 9(4):490–495
Wu H, Neale MC (2013) On the likelihood ratio tests in bivariate ACDE models. Psychometrika 78(3):441–463
Acknowledgements
This study was supported by National Institutes of Health (Grant No. DA030005, DA025109, DA018673, DA024304, U01 DA04112).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
All authors reviewed and approved the final manuscript before its submission.
Human and Animal Rights and Informed consent
Ethical approval for the study was obtained from the Institutional Review Board of Virginia Commonwealth University. All participants provided informed consent prior to participating in the study.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Edited by Meike Bartels.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Maes, H.H., Neale, M.C., Kirkpatrick, R.M. et al. Using Multimodel Inference/Model Averaging to Model Causes of Covariation Between Variables in Twins. Behav Genet 51, 82–96 (2021). https://doi.org/10.1007/s10519-020-10026-8
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10519-020-10026-8