Abstract
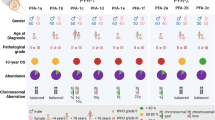
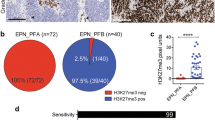
The PFA molecular subgroup of posterior fossa ependymomas (PF-EPNs) shows poor outcome. H3K27me3 (me3) loss by immunohistochemistry (IHC) is a surrogate marker for PFA wherein its loss is attributed to overexpression of Cxorf67/EZH2 inhibitory protein (EZHIP), C17orf96, and ATRX loss. We aimed to subgroup PF-EPNs using me3 IHC and study correlations of the molecular subgroups with other histone related proteins, 1q gain, Tenascin C and outcome. IHC for me3, acetyl-H3K27, H3K27M, ATRX, EZH2, EZHIP, C17orf96, Tenascin-C, and fluorescence in-situ hybridisation for chromosome 1q25 locus were performed on an ambispective PF-EPN cohort (2003–2019). H3K27M-mutant gliomas were included for comparison. Among 69 patients, PFA (me3 loss) constituted 64%. EZHIP overexpression and 1q gain were exclusive to PFA seen in 72% and 19%, respectively. Tenascin C was more frequently positive in PFA (p = 0.02). H3K27M expression and ATRX loss were noted in one case of PFA–EPN each. All H3K27M-mutant gliomas (n = 8) and PFA-EPN (n = 1) were EZHIP negative. C17orf96 and acetyl-H3K27 expression did not correlate with me3 loss. H3K27me3 is a robust surrogate for PF-EPN molecular subgrouping. EZHIP overexpression was exclusive to PFA EPNs and was characteristically absent in midline gliomas and the rare PFA harbouring H3K27M mutations representing mutually exclusive pathways leading to me3 loss.



Similar content being viewed by others
Change history
09 January 2021
In the original publication of the article, the middle name was missing in corresponding author’s name.
References
Ellison DW, McLendon R, Wiestler OD et al (2016) Ependymoma. In: Louis DN, Ohgaki H, Wiestler OD, Cavenee WK (eds) WHO classification of tumours of the central nervous system, revised, 4th edn. IARC, Lyon, pp 106–112
Dyer S, Prebble E, Davison V et al (2002) Genomic imbalances in pediatric intracranial ependymomas define clinically relevant groups. Am J Pathol 161:2133–2141
Pajtler KW, Mack SC, Ramaswamy V et al (2017) The current consensus on the clinical management of intracranial ependymoma and its distinct molecular variants. Acta Neuropathol 133:5–12
Korshunov A, Witt H, Hielscher T et al (2010) Molecular staging of intracranial ependymoma in children and adults. J Clin Oncol 28:3182–3190
Witt H, Mack SC, Ryzhova M et al (2011) Delineation of two clinically and molecularly distinct subgroups of posterior fossa ependymoma. Cancer Cell 20:143–157
Wani K, Armstrong TS, Vera-Bolanos E et al (2012) A prognostic gene expression signature in infratentorial ependymoma. Acta Neuropathol 123:727–738
Mack SC, Witt H, Piro RM, Gu L, Zuyderduyn S, Stütz AM et al (2014) Epigenomic alterations define lethal CIMP-positive ependymomas of infancy. Nature 506:445–450
Bayliss J, Mukherjee P, Lu C et al (2016) Lowered H3K27me3 and DNA hypomethylation define poorly prognostic pediatric posterior fossa ependymomas. Sci Transl Med. 8:366ra161
Pajtler KW, Witt H, Sill M, Jones DT, Hovestadt V, Kratochwil F et al (2015) Molecular classification of ependymal tumors across all CNS compartments, histopathological grades, and age groups. Cancer Cell 27:728–743
Pajtler KW, Wen J, Sill M et al (2018) Molecular heterogeneity and CXorf67 alterations in posterior fossa group A (PFA) ependymomas. Acta Neuropathol 136(2):211–226
Ramaswamy V, Hielscher T, Mack SC et al (2016) Therapeutic impact of cytoreductive surgery and irradiation of posterior fossa ependymoma in the molecular era: a retrospective multicohort analysis. J Clin Oncol 34:2468–2477
Louis DN, Wesseling P, Aldape K et al (2020) cIMPACT-NOW update 6: new entity and diagnostic principle recommendations of the cIMPACT-Utrecht meeting on future CNS tumor classification and grading. Brain Pathol. https://doi.org/10.1111/bpa.12832 (published online ahead of print, 2020 Apr 19)
Zapotocky M, Beera K, Adamski J et al (2019) Survival and functional outcomes of molecularly defined childhood posterior fossa ependymoma: cure at a cost. Cancer 125:1867–1876
Nambirajan A, Malgulwar PB, Sharma A et al (2019) Clinicopathological evaluation of PD-L1 expression and cytotoxic T-lymphocyte infiltrates across intracranial molecular subgroups of ependymomas: are these tumors potential candidates for immune check-point blockade? Brain Tumor Pathol 36:152–161
Michealraj KA, Kumar SA, Kim LJY et al (2020) Metabolic regulation of the epigenome drives lethal infantile ependymoma. Cell 181:1329–1345
Hübner JM, Müller T, Papageorgiou DN et al (2019) EZHIP/CXorf67 mimics K27M mutated oncohistones and functions as an intrinsic inhibitor of PRC2 function in aggressive posterior fossa ependymoma. Neuro Oncol 21:878–889
Jain SU, Do TJ, Lund PJ et al (2019) PFA ependymoma-associated protein EZHIP inhibits PRC2 activity through a H3 K27M-like mechanism. Nat Commun 10:2146
Beringer M, Pisano P, Di Carlo V et al (2016) EPOP functionally links elongin and polycomb in pluripotent stem cells. Mol Cell 64:645–658
Castel D, Philippe C, Calmon R et al (2015) Histone H3F3A and HIST1H3B K27M mutations define two subgroups of diffuse intrinsic pontine gliomas with different prognosis and phenotypes. Acta Neuropathol 130:815–827
Panwalkar P, Clark J, Ramaswamy V, Hawes D, Yang F, Dunham C et al (2017) Immunohistochemical analysis of H3K27me3 demonstrates global reduction in group-A childhood posterior fossa ependymoma and is a powerful predictor of outcome. Acta Neuropathol 134:705–714
Fukuoka K, Kanemura Y, Shofuda T et al (2018) Significance of molecular classification of ependymomas: C11orf95-RELA fusion-negative supratentorial ependymomas are a heterogeneous group of tumors. Acta Neuropathol Commun 6:134
Tanrıkulu B, Danyeli AE, Özek MM (2020) Is H3K27me3 status really a strong prognostic indicator for pediatric posterior fossa ependymomas? A single surgeon, single center experience. Childs Nerv Syst 36:941–949
Ellison DW, Kocak M, Figarella-Branger D et al (2011) Histopathological grading of pediatric ependymoma: reproducibility and clinical relevance in European trial cohorts. J Negat Results Biomed 10:7
Rajeshwari M, Sharma MC, Kakkar A et al (2016) Evaluation of chromosome 1q gain in intracranial ependymomas. J Neurooncol 127:271–278
Jha P, Manjunath N, Singh J et al (2019) Analysis of PD-L1 expression and T cell infiltration in different molecular subgroups of diffuse midline gliomas. Neuropathology 39:413–424
Andreiuolo F, Le Teuff G, Bayar MA et al (2017) Integrating Tenascin-C protein expression and 1q25 copy number status in pediatric intracranial ependymoma prognostication: a new model for risk stratification. PLoS ONE 12:e0178351
Upadhyaya SA, Robinson GW, Onar-Thomas A et al (2019) Molecular grouping and outcomes of young children with newly diagnosed ependymoma treated on the multi-institutional SJYC07 trial. Neuro Oncol 21:1319–1330
Gessi M, Capper D, Sahm F et al (2016) Evidence of H3 K27M mutations in posterior fossa ependymomas. Acta Neuropathol 132:635–637
Ryall S, Guzman M, Elbabaa SK et al (2017) H3 K27M mutations are extremely rare in posterior fossa group A ependymoma. Childs Nerv Syst 33:1047–1051
Weinberg DN, Allis CD, Lu C (2017) Oncogenic mechanisms of histone H3 mutations. Cold Spring Harb Perspect Med 7:a026443
Rogers HA, Chapman R, Kings H et al (2018) Limitations of current in vitro models for testing the clinical potential of epigenetic inhibitors for treatment of pediatric ependymoma. Oncotarget 9:36530–36541
Li AM, Dunham C, Tabori U et al (2015) EZH2 expression is a prognostic factor in childhood intracranial ependymoma: a Canadian Pediatric Brain Tumor Consortium study. Cancer 121:1499–1507
Venneti S, Garimella MT, Sullivan LM et al (2013) Evaluation of histone 3 lysine 27 trimethylation (H3K27me3) and enhancer of Zest 2 (EZH2) in pediatric glial and glioneuronal tumors shows decreased H3K27me3 in H3F3A K27M mutant glioblastomas. Brain Pathol 23:558–564
Acknowledgements
This work is supported by Central Institute of Industrial research (CSIR), New Delhi, India (Pool No 8948A/17), and Neurosciences centre, AIIMS, New Delhi.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
All authors declare no conflicts of interest.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
The original online version of this article was revised due to the name of corresponding author was published incorrectly and corrected in this version.
Electronic supplementary material
Below is the link to the electronic supplementary material.
10014_2020_385_MOESM1_ESM.tif
Supplementary Figure 1: Tukey plot showing EZH2 scores among different subgroups of PF-EPNs and H3K27M-mutant brain stem DMG in the study. *Significant difference in score was seen only between EZHIP positive and negative PF-EPNs and none of the other sub groups (TIF 17 kb)
10014_2020_385_MOESM2_ESM.tif
Supplementary Figure 2: Histomorphology and immunohistochemical features of the H3K27M-mutant PFA ependymoma. The histomorphology was that of a classical Grade 2 ependymoma with perivascular pseudo-rosettes (A) and showed characteristic dot-like staining for epithelial membrane antigen (B). There was loss of staining for H3K27me3 in tumour cells (C) with strong diffuse staining for H3K27M protein (D). Sanger sequencing chromatogram shows presence of H3F3A p.K27M (orange arrows) in tumour cells using reverse (E) and forward (F) primers (TIF 4016 kb)
10014_2020_385_MOESM3_ESM.tif
Supplementary Figure 3: Histomorphology and immunohistochemical features of H3K27M-mutant diffuse midline gliomas. A representative case showing histomorphology of an infiltrating glioma (A) showing loss of H3K27me3 staining in tumour cells (B) with strong diffuse staining for H3K27M protein (D). EZH2 expression was variable (E) while there was diffuse staining for acetyl-H3K27 (F) (TIF 6007 kb)
Rights and permissions
About this article
Cite this article
Nambirajan, A., Sharma, A., Rajeshwari, M. et al. EZH2 inhibitory protein (EZHIP/Cxorf67) expression correlates strongly with H3K27me3 loss in posterior fossa ependymomas and is mutually exclusive with H3K27M mutations. Brain Tumor Pathol 38, 30–40 (2021). https://doi.org/10.1007/s10014-020-00385-9
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10014-020-00385-9