Abstract
Parameter estimation problems of mathematical models can often be formulated as nonlinear least squares problems. Typically these problems are solved numerically using iterative methods. The local minimiser obtained using these iterative methods usually depends on the choice of the initial iterate. Thus, the estimated parameter and subsequent analyses using it depend on the choice of the initial iterate. One way to reduce the analysis bias due to the choice of the initial iterate is to repeat the algorithm from multiple initial iterates (i.e. use a multi-start method). However, the procedure can be computationally intensive and is not always used in practice. To overcome this problem, we propose the Cluster Gauss–Newton (CGN) method, an efficient algorithm for finding multiple approximate minimisers of nonlinear-least squares problems. CGN simultaneously solves the nonlinear least squares problem from multiple initial iterates. Then, CGN iteratively improves the approximations from these initial iterates similarly to the Gauss–Newton method. However, it uses a global linear approximation instead of the Jacobian. The global linear approximations are computed collectively among all the iterates to minimise the computational cost associated with the evaluation of the mathematical model. We use physiologically based pharmacokinetic (PBPK) models used in pharmaceutical drug development to demonstrate its use and show that CGN is computationally more efficient and more robust against local minima compared to the standard Levenberg–Marquardt method, as well as state-of-the art multi-start and derivative-free methods.
Similar content being viewed by others
1 Introduction
The parameter estimation of mathematical models often boils down to solving nonlinear least squares problems. Hence, algorithms for solving nonlinear least squares problems are widely used in many scientific fields.
The most traditional least squares solver is the Gauss–Newton method (Gauss 1857; Björck 1996). In practice, the Gauss–Newton method with regularisation [i.e., Levenberg–Marquardt (LM) method Marquardt 1963; Moré 1978] or with the Trust-Region method (Conn et al. 2000) is often used. Recently, derivative-free methods, which do not explicitly use derivative information of the nonlinear function, have been developed. These methods are usually computationally more efficient as it avoids the costly computation of the derivatives of the nonlinear functions. Also, they can be applied even to problems where the mathematical models are ‘black box’. The state of the art derivative-free algorithms are DFO-LS (Cartis and Roberts 2019) and POUNDERS (Wild 2017). A comprehensive review of the derivative-free methods can be found in Larson et al. (2019).
Another approach for obtaining a solution of nonlinear least squares problems is to directly minimise the sum of squared residuals (SSR) using generic optimisation algorithms for the scalar objective function. The most classical approach is to obtain a minimiser where the gradient of the SSR becomes zero using the Newton method. As it is usually too costly to compute the Hessian of the SSR, Quasi-Newton methods which approximate the Hessian are used. The commonly used Quasi-Newton method is the BFGS method (Broyden 1970; Fletcher 1970; Goldfarb 1970; Shanno 1970; Shanno and Kettler 1970). Another approach which makes use of the Newton-type method for optimisation is Implicit Filtering (Kelley 2011) which combines grid search and the Newton method. In addition to these optimisation algorithms, we can use numerous global optimisation algorithms when bound constraints are given. For example, Surrogate Optimisation (Gutmann 2001), Genetic Algorithm (Goldberg and Holland 1988), Particle Swarm algorithm (Kennedy and Eberhart 1995; Mezura-Montes and Coello 2011), and DIRECT (Jones et al. 1993) are well known global optimisation algorithms.
Although there are a variety of algorithms to solve the nonlinear least squares problems as listed above, they mostly focus on finding one minimiser. To the best of our knowledge, there is very limited methodological development on algorithms for simultaneously finding multiple approximate minimisers of nonlinear least squares problems. For instance, when using the Levenberg–Marquardt method, the local algorithm often gives a local minimiser which depends on the choice of the initial iterate. To reduce the analysis bias due to the initial iterate used for a local algorithm, it is a good practice to repeatedly use the local algorithm with various initial iterates, as in multi-start methods (Boender et al. 1982). Similarly, for problems where bound constraints are given, one can use global optimisation algorithms to find one of the global minimisers. On the other hand, if there are multiple global minisers, the global minimiser found can depend on the algorithm setting, for example, the random seed. Hence, it is beneficial to use global optimisation algorithms with various settings repeatedly if the uniqueness of the global minimiser is not guaranteed. The trivial bottleneck of repeatedly using these algorithms is the computation cost. In this paper, we propose a new method addressing this computational challenge of finding multiple approximate minimisers of nonlinear least squares problems.
Our algorithm development for finding multiple local minimisers of nonlinear least squares problems was motivated by a mathematical model of pharmaceutical drug concentration in a human body called the physiologically based pharmacokinetic (PBPK) model (Watanabe et al. 2009). The PBPK model is typically a system of mildly nonlinear stiff ordinary differential equations (ODEs) with many parameters. This type of mathematical model is constructed based on the knowledge of the mechanism of how the drug is absorbed, distributed, metabolised and excreted. Given the complexity of this process and the limitation of the observations we can obtain from a live human subject, the model parameters cannot be uniquely identified from the observations, meaning that there are non-unique global minimisers to the nonlinear least squares problem. The estimated parameters of the PBPK model are used to simulate the drug concentration of the patient from whom we are often unable to test the drug on (e.g., children, pregnant person, a person with rare genetic anomaly) or to predict the experiment that is yet to be run (different amount of drug administration, multiple drugs used at the same time). As the simulated drug concentration is used to predict the safety of the drug in these different scenarios, it is essential to consider multiple predictions based on multiple possible parameters that are estimated from the available observations. A motivating example is presented in Sect. 3. Another reason why we want to obtain multiple sets of parameters is that we can understand which parameters cannot be estimated from the available data. This will motivate the pharmaceutical scientists to perform additional (e.g., in-vitro or in-animal) experiments to determine these parameters that were not estimable from the available data.
In Aoki et al. (2011) and Aoki et al. (2014) we proposed the Cluster Newton (CN) method for obtaining multiple solutions of a system of underdetermined nonlinear equations. In recent years CN has been used in the field of pharmaceutical science (Yoshida et al. 2013; Fukuchi et al. 2017; Asami et al. 2017; Toshimoto et al. 2017; Kim et al. 2017; Nakamura et al. 2018). For example, Toshimoto et al. (2017) used the parameters estimated by CN to predict the adverse drug effect, Nakamura et al. (2018) used the estimated parameters to predict the outcome of a clinical trial. However, based on these applications of CN, we observed the necessity to develop a new algorithm for finding multiple approximate minimisers of a nonlinear least squares problem which is more robust against noise in the observed data. This is mainly because actual pharmaceutical data may contain measurement error and inconsistency coming from an inadequate model.
1.1 Nonlinear least squares problem of our interest
In this paper, we propose an algorithm for obtaining multiple approximate minimisers of nonlinear least squares problems
which do not have a unique solution (global minimiser), that is to say, there exist \(\varvec{x}^{(1)}\ne \varvec{x}^{(2)}\) such that
Here, \(\varvec{f}\) is a nonlinear function from \(\mathbb {R}^n\) to \(\mathbb {R}^m\), \(\varvec{x}^{(1)},\,\varvec{x}^{(2)}\in \mathbb {R}^n\) and \(\varvec{y}^*\in \mathbb {R}^m\). The nonlinear function \(\varvec{f}\) can be derived from a mathematical model, the vector \(\varvec{x}\) can be regarded as a set of model parameters which one wishes to estimate, and the vector \(\varvec{y}^*\) can be regarded as a set of observations one wishes to fit the model to. In the motivating problem of estimating parameters of PBPK models, since there are usually insufficient observations, the corresponding nonlinear least squares problem has multiple global minimisers. Therefore, we are interested in finding multiple minimisers instead of just one minimiser.
We shall call the following quantity the sum of squared residuals (SSR):
and use it for the quantification of the goodness of \(\varvec{x}\) as the approximation of the solution of the least squares problem (1).
1.2 Well known example in pharmacokinetics
In this subsection, we present a simple pharmacokinetics parameter estimation problems where the corresponding nonlinear least squares problems have non-unique global minimisers. This example is called ‘flip-flop kinetics’ and can be found in most standard textbooks in pharmacokinetics (e.g. Gibaldi and Perrier 1982). Flip-flop kinetics occurs when estimating the pharmacokinetic parameters of the drug that is orally given (e.g., as a pill or a tablet) to patients based on the observation of the drug concentration in the blood plasma. The simplest mathematical model for this pharmacokinetics can be written as follows:
where
For this problem \(u_2\) corresponds to the drug concentration in the blood plasma, which is observable (i.e., \(u_2(t_i)\) is the model simulation corresponding to the observation \(y^*_i\), where \(t_i\) is the ith observation time.). It can be shown analytically that there are two distinct parameter sets that realise the same drug concentration time-course curve. Figure 1 shows the surface plot of the sum of squared residuals of the corresponding nonlinear least squares problem. As can be seen, there are two global minimisers for this nonlinear least squares problem.
In this subsection, we have shown the simplest possible form of this issue of non-unique global minimisers in pharmacokinetics. Thus, we would like to point out that one cannot assume the uniqueness of the global minimiser for more complex pharmacokinetic models, for example, the PBPK model of our interest.
2 Method: Algorithm
In this section we describe the proposed algorithm. We first introduce a rough concept using a toy example in Sect. 2.1 and then introduce the full algorithm in detail in Sect. 2.2.
2.1 Brief explanation of the algorithm
The aim of the proposed Cluster Gauss–Newton (CGN) algorithm is to efficiently find multiple approximate minimisers of the nonlinear least squares problem (1). We do so by first creating a collection of initial guesses which we call the ‘cluster’. Then, we move the cluster iteratively using linear approximations of the nonlinear function \(\varvec{f}\), similarly to the Gauss–Newton method (Björck 1996).
The unique idea in the CGN method is that the linear approximation is constructed collectively throughout the points in the cluster instead of using the Jacobian matrix which approximates the nonlinear function linearly at a point as in the Gauss–Newton or LM. By using points in the cluster to construct a linear approximation, instead of explicitly approximating the Jacobian, we minimise the computational cost associated with the nonlinear function (i.e., mathematical model) for each iteration. In addition, by constructing linear approximation using non-local information, CGN is more likely to converge to approximate minimisers with smaller SSR than methods using the Jacobian.
In order to visualise the key differences between the proposed linear approximation (CGN) and the Jacobian (i.e., derivative) approaches, we consider the nonlinear function
(see Fig. 2) and aim to find global minimisers. Any point \(x\in [-1,1]\) is a global minimiser of this problem. Hence, this problem has nonunique global minimisers. Let the points of the initial iterates be:
We now compute the linear approximations used to move these points in the cluster to minimise the function f.
Gradient (LM)
For this nonlinear function, since the function is given in analytic form, we can obtain the gradient explicitly. In, practice, when f is given as a “black box”, we can approximate the derivative by a finite difference scheme, for example, \(f'(x_i)\approx \frac{f(x_i+\epsilon )-f(x_i)}{\epsilon }\). Then, the linear approximation at \(x_i\) can be written as \(f(x) \approx \frac{f(x_i+\epsilon )-f(x_i)}{\epsilon }(x-x_i)+f(x_i)\). Notice that it requires one extra evaluation of f at \(x_i+\epsilon\) for each \(x_i\). This number of extra function evaluation is, when evaluating a full gradient estimate, equal to the number of independent variables of f. (This is not the case if a directional derivative estimate is used.) If f is given by a system of ODEs, one may use the adjoint method to obtain the derivatives more efficiently. However, it requires solving an additional system of ODEs (the adjoint equation). More importantly, iterates of methods based on the gradient may converge to local minimisers, since they use local gradient information.
Cluster Gauss–Newton (CGN) method (proposed method)
In the proposed method, we construct a linear approximation for each point in the cluster while using the value of f at other points in the cluster to globally approximate the nonlinear function with a linear function. The influence of another point in the cluster to the linear approximation is weighted according to how close the point is to the point of approximation, i.e.,
where \(a_{(i)}\) is the slope of the linear approximation at \(x_i\) and the linear approximation at \(x_i\) can be written as \(f(x)\approx a_{(i)}(x-x_i)+f(x_i)\). There are many possibilities for the weight \(d_{j(i)}\). In this paper, we choose \(d_{j(i)}=(x_j-x_i)^{-2\gamma }\) where \(\gamma \ge 0\) (\(\gamma = 0\) corresponds to uniform weight). Note that Eq. (9) can also be regarded as a weighted least squares solution of a system of linear equations where the weight is chosen to be \(d_{j(i)}\). The weight is motivated by the fact that we weight the information from the neighbouring points in the cluster more than the ones further away when constructing the linear approximation. Note that we do not require any extra evaluation of f for obtaining these linear approximations.
For the multi-dimensional nonlinear function \(\varvec{f}:\mathbb {R}^n\rightarrow \mathbb {R}^m\), when we have N points in the cluster, we solve the following linear least squares problem:
where \(D_{(i)}=\text {diag}(d_{1i}, \ldots ,d_{Ni})\) is a diagonal matrix defining the weights, \(\varDelta X_{(i)}\in \mathbb {R}^{N\times n}\) is the difference between all the cluster points and \(\varvec{x}_i\), and \(\varDelta Y_{(i)}\in \mathbb {R}^{N\times m}\) is the difference between the nonlinear function \(\varvec{f}\) evaluated at all the cluster points and at \(\varvec{x}_i\). The precise definition of these quantities and derivation of (10) is given in the next subsection.
For the one dimensional case (i.e., \(m=n=1\)), the linear approximations at each point for the first ten iterations for both CGN and LM are shown in Fig. 2. As can be seen, the gradient used in LM captures the local behaviour of the nonlinear function. The linear approximation used in CGN, on the other hand, captures the global behaviour of the nonlinear function. After nine iterations of the CGN, all the points reached the minimisers with smallest SSR. On the other hand, the LM converges to local minimisers whose SSR are not necessarily the minimum.
Schematic comparison of CGN and LM. Dots represent the iterates with their function values in each iteration and the lines represent linear approximations used to update the iterates. As can be seen in this figure, the linear approximation used in CGN follows the global trend of the function, while the slope used in LM captures the local feature of the function. As a result, when using CGN all the iterates converge to the minimisers with the smallest residual (in this case global minimisers), while all the iterates of LM converge to local minimisere whose residuals are not the minimum. In addition, for the 10 iterations presented in the figure, CGN required only 50 function evaluations while LM required 121 function evaluations since the slope is approximated using finite differences
2.2 Detailed description of the algorithm
Next, we describe the proposed CGN algorithm in detail. In this subsection, we denote a scalar quantity by a lower case letter e.g., a, c, a matrix by a capital letter, e.g., A, or M, and a column vector by a bold symbol of a lower case letter, e.g., \(\varvec{v}\), \(\varvec{a}\), unless otherwise specifically stated. Super script \(\text {T}\) indicates the transpose. Hence, \(\varvec{v}^{\text {T}}\) and \(\varvec{a}^{\text {T}}\) are row vectors.
(1) Pre-iteration process
(1-1) Create initial cluster
The initial iterates of CGN, a set of vectors \(\{\varvec{x}_{i}^{(0)}\}_{i=1}^{N}\) are genrated using uniform random numbers in each component within the domain of initial estimate of the plausible location of global minimisers given by the user. The unique point of the CGN is that the user specifies the domain of the initial estimates instead of a point. In this paper, we assume that the domain of initial guess is given by the user by two sets of vectors \(\varvec{x}^\text {L}\), \(\varvec{x}^\text {U}\), and the value \(x_{ji}^{(0)}\) is sampled from the uniform distribution between \(x^\text {L}_j\) and \(x^\text {U}_j\), where \(x_{ji}^{(0)}\) is the j th element of vector \(\varvec{x}_i^{(0)}\).
Note that this does not mean that all the following iterates \(\varvec{x}_i^{(k)} \quad (k \ge 1, i=1,2, \ldots , N)\) must satisfy \(\varvec{x}^L \le \varvec{x}_i^{(k)} \le \varvec{x}^U\). Also note that here we have used uniform distribution; however, other choices of initial distributions are possible. A brief numerical experiments using different initial distributions can be found in “Appendix C”.
Store the initial set of vectors in a matrix \(X^{(0)}\), i.e.,
where the super script (0) indicates the initial iterate.
Evaluate the nonlinear function \(\varvec{f}\) at each \(\varvec{x}_{i}^{(0)}\) as \(\varvec{y}_{i}^{(0)}=\varvec{f}(\varvec{x}_{i}^{(0)}) \quad (i=1,2,\ldots ,N)\) and store in matrix \(Y^{(0)}\), i.e.,
If the function \(\varvec{f}\) cannot be evaluated at \(\varvec{x}_{i}^{(0)}\), then re-sample \(\varvec{x}_{i}^{(0)}\) until \(\varvec{f}\) can be evaluated.
Compute the sum of squared residuals vector \(\varvec{r}^{(0)}\) i.e.,
The concise pseudo-code for the creation of the initial cluster can be found in Algorithm 1.

(1-2) Initialise regularisation parameter vector
Fill the regularisation parameter vector \(\varvec{\lambda }^{(0)}\in \mathbb {R}^N\), with the user-specified initial regularisation parameter \(\lambda _\text {init}\) .i.e.,
(2) Main iteration
Repeat the following procedure until the user specified stopping criteria are met. We denote the iteration number as k, which starts from 0 and is incremented by 1 after each iteration.
(2-1) Construct weighted linear approximations of the nonlinear function
We first construct a linear approximation around the point \(\varvec{x}_{i}^{(k)}\), s.t.,
Here, \(A^{(k)}_{(i)} \in \mathbb {R}^{m\times n}\) describes the slope of the linear approximation around \(\varvec{x}_{i}^{(k)}\).
The key difference of our algorithm compared to others is that we construct a Jacobian like matrix \(A_{(i)}^{(k)}\) collectively using all the function evaluations of \(\varvec{f}\) in the previous iteration, i.e., we solve
for \(i=1,...,N\), where \(d_{j(i)}^{(k)}\ge 0\), \(j=1,...,N\) are weights. Here, \(\varDelta \varvec{y}_{j(i)}^{(k)} = \varvec{f}(\varvec{x}_j^{(k)})-\varvec{f}(\varvec{x}_i^{(k)})\in \mathbb {R}^m\) and \(\varDelta \varvec{x}_{j(i)}^{(k)} = \varvec{x}_j^{(k)}-\varvec{x}_i^{(k)}\in \mathbb {R}^n\). (Note that \(\varDelta \varvec{y}_{i(i)}^{(k)} = \varvec{0}\), \(\varDelta \varvec{x}_{i(i)}^{(k)} = \varvec{0}\).) Also, let
Note that \(\varvec{f}(\varvec{x}_{i}^{(k)})\) are always computed at the previous iteration [e.g., as Eq. (12) when \(k=0\) and in Step 2–3 when \(k>0\)]. Hence, no new evaluation of \(\varvec{f}\) is required at this step.
The key idea here is that we weight the information of the function evaluation near \(\varvec{x}_{i}^{(k)}\) more than the function evaluation further away. That is to say, \(d_{j(i)}^{(k)}>d_{j'(i)}^{(k)}\) if \(||\varvec{x}_{j}^{(k)}-\varvec{x}_{i}^{(k)}||<||\varvec{x}_{j'}^{(k)}-\varvec{x}_{i}^{(k)}||\). The importance of this idea can be seen in the numerical experiment presented in “Appendix A”.
Noting that
we can rewrite Eq. (17) as
where
where \(d^{(k)}_{l(i)}\ge 0\). In this paper, we choose the weights as
where \({x}_{lj}^{(k)}, x_l^\text {U}, x_l^\text {L}\) are the l th element of the vectors \(\varvec{x}_{j}^{(k)}, \varvec{x}^\text {U}, \varvec{x}^\text {L}\), respectively (\(l=1,...,n\)), and \(\gamma \ge 0\) is a constant. We use this weighting scheme so that the “information” from the nonlinear function evaluation from the point closer to the point of approximation is more influential when constructing the linear approximation. The distance between \(\varvec{x}_{i}\) and \(\varvec{x}_{j}\) are normalised by the size of the domain of initial guess (i.e., \(\varvec{x}^\text {U}\) and \(\varvec{x}^\text {L}\)). The effect of the weight \(d_{j(i)}^{(k)}\) and the parameter \(\gamma\) and its necessity is analysed in “Appendix A”. The minimum norm solution of Eq. (21) is given by
where \(^\dagger\) denotes the Moore–Penrose inverse.
If \({\mathrm{rank}} \varDelta X\,{(k)}_{(i)} D^{(k)}_{(i)} = n\),
Generically, \(\text {rank} \varDelta X^{(k)}_{(i)} D^{(k)}_{(i)} = n.\) \(\text {rank} \varDelta X^{(k)}_{(i)} < n\) can happen when \(x^{(k)}_{li} = c \quad (i=1,2,\ldots , N)\), i.e. when \(\varvec{x}^{(k)}_{i}\) lie in the same hyperplane \(x_l = c\). This happens when the l-th component of \(\varvec{x}^{(k)}_i \quad (i=1,2,\ldots ,N)\) are all equal.
The concise pseudo-code for the weighted linear approximation can be found in Algorithm 2.

(2-2) Solve for \(\varvec{x}\) that minimises \(||\varvec{y}^*- (A_{(i)}^{(k)} (\varvec{x}-\varvec{x}_{i}^{(k)})+\varvec{f}(\varvec{x}_{i}^{(k)}))||^{\,2}_2\)
We now compute the next iterate \(X^{(k+1)}\) using the matrices \(\{A^{(k)}_{(i)}\}_{i=1}^{N}\) similarly to the Gauss–Newton method with Tikhonov regularisation (e.g., Hansen 2005; Björck 1996), i.e.,
for \(i=1,...,N\), where \(\varvec{y}^*\) is the set of observations one wishes to fit the nonlinear function \(\varvec{f}\) to [cf. (1)], and \(\varvec{y}_i^{(k)}\equiv \varvec{f}(\varvec{x}_i^{(k)})\). For CGN, we require \(\lambda _i^{(k)}> 0\). The necessity of the regularisation can be seen in the numerical experiment presented in “Appendix B”.
(2-3) Update matrices X and Y and vectors \(\varvec{r}\) and \(\varvec{\lambda }\)
Evaluate the nonlinear function \(\varvec{f}\) for each \(\varvec{x}_{i}^{(k+1)}\) as \(\varvec{y}_i^{(k+1)} = \varvec{f}(\varvec{x}_i^{(k+1)} )\) \((i=1,2,\ldots ,n)\), and store as \(Y^{(k+1)}= [ {y}_1^{(k+1)}, \varvec{y}_2^{(k+1)},\ldots ,\varvec{y}_N^{(k+1)} ].\) Compute the sum of squared residuals vector as \(\varvec{r}^{(k+1)}= [ r_1^{(k+1)}, r_2^{(k+1)},\) \(\ldots , r_N^{(k+1)} ]^\text {T}\) where \(r_i^{(k+1)} = \Vert {y}_i^{(k+1)} - {y}^{*} \Vert _2^{\,2} \quad (i=1,2,\ldots ,N)\). Note that this process can be implemented in an embarrassingly parallel way.
If the residual \(r_i\) increases, we replace \(\varvec{x}_{i}^{(k+1)}\) by \(\varvec{x}_{i}^{(k)}\) and increase the regularisation parameter i.e.,
if \(r_{i}^{(k)}<r_{i}^{(k+1)}\) or \(\varvec{f}(\varvec{x}_{i}^{(k+1)})\) can not be evaluated, then let
else decrease the regularisation parameter, i.e.,
for each \(i=1,..., N\).
There are various ways to update the regularisation parameter \(\lambda\). In this paper we followed the strategy used in the Matlab’s implementation of the Levenberg–Marquardt method.
In addition, in this step, we impose the stopping criteria for each point in the iteration. As can be seen in Eq. (26), \(\varvec{x}_{i}^{(k+1)}\approx \varvec{x}_{i}^{(k)}\) for large \(\lambda _i\), so that we can expect very small update in \(\varvec{x}_{i}^{(k+1)}\). Hence, in order to mimic the minimum step size stopping criteria, we stop the update for i where \(\lambda _i>\lambda _\text {max}\).
The concise pseudo-code for updating matrices X and Y and vectors \(\varvec{r}\) and \(\varvec{\lambda }\) is given in Algorithm 3.
The influence of the choice of the initial value \(\lambda _{\text {init}}\) of the regularization parameter in Eq. (15) is studied in “Appendix B”.

We present a concise description of the CGN as a pseudo-code in Algorithm 4.

3 Motivating example
In this section, we introduce a motivating example to illustrate how the proposed method could be used in practice. We consider a scenario where a newly developed drug is tested for the first time in a human. Before the drug is given to a human, the biochemical properties of the drug are studied in-test-tube (in-vitro) and in-animal experiments. However, how the drug behaves in the human body is still uncertain. Based on the results of in-vitro and in-animal experiments, the team decides that 100mg is a safe amount of the drug to be given to a human and the experiment is conducted with a healthy normal volunteer, and the drug concentration in the blood plasma is measured at points in time. Using these measurements, we estimate the multiple possible model parameters which can be used to simulate various scenarios. The following workflow can be envisioned:
1: Construct a mathematical model based on the understanding of the physiology and biochemical properties of the drug.
In this example, we use the model presented in Watanabe et al. (2009). The mathematical model is depicted in Fig. 3, and it can be written as a system of nonlinear ordinary differential equations with 20 variables. We refer the readers to the supplementary material for the complete description of the model. There are two types of model parameters in this model: physiological parameters and kinetic parameters. Examples of the physiological parameters are the sizes of the organs or the blood flow rates between the organs. As the human physiology is well studied and these parameters usually do not depend on the drugs, we can assume these parameters to be known. The kinetic parameters, such as, how fast the drug gets excreted from the body or how easily it binds to tissues are the parameters that depend on the drug and usually are not very well known. Before the first in-human experiment, the drug development team characterises these parameters using an organ in test-tubes or by administering the drug to an animal. However, these parameters can differ from animal to human, so we do not have a very accurate estimate of these parameters. The differences in these parameters between a human and an animal can be several orders of magnitude. Figure 4 depicts plots of the drug concentration simulation where the kinetic parameters are sampled within a reasonable range of the parameters. As can be seen in Fig. 4, we cannot obtain any useful information just by randomly sampling the kinetic parameters from the feasible range.
A schematic diagram of a physiologically based pharmacokinetic model. (Arrows represent the movement of the drug to a different part of the body. Variables \(u_i\) are the drug concentration or the amount of the drug in each compartment. The body is divided into Blood, Muscle, Skin, Adipose, Liver and Intestine. The liver is further divided into ten compartments to model the complex drug behaviour in the liver. The intestine is divided into four compartments, three transit compartments and one intestine compartment, to model the time it takes for the drug to reach the intestine)
2: Sample multiple possible model parameter sets that fit the model prediction of the drug concentration to the observed data from the 100mg experiment. We now use the observed data from the experiment where 100mg of the drug was given to a human. The red dots in Fig. 5 depict the observed data. The left panel of Fig. 5 shows some of the simulation results using the parameter sets of the initial iterate of CGN. The right panel of Fig. 5 shows some of the simulation results after 20 iterations of CGN. As can be seen in Fig. 5, CGN can find multiple sets of parameters that fit the observed data. The parameter values are depicted in the box plots in Fig. 6. As can be seen in Fig. 6, after 20 iterations of CGN, the distribution of some of the parameters shrinks significantly suggesting these parameters can be identified from the observations while the distribution of some of the parameters are unchanged indicating that these parameters cannot be identified from the observation. In Fig. 7, we show scatter plots of the parameters found by CGN. As can be seen in Fig. 7, even if the parameter cannot be identified from the observation, some nonlinear relationships can occasionally be identified between the parameters.
Plot of the simulation of drug concentration (black solid line) with observations (red dot). Simulation results are based on the parameters for the initial iterate for CGN and the parameters found after 20 iterations of CGN. In the left panel, the simulation results based on the top 100 sets of the parameters (out of 1000 parameter sets in the cluster) from the initial cluster are shown. In the right panel, the simulations results based on the top 100 sets of the parameters (out of 1000 parameter sets in the cluster) after 20 iterations of CGN are shown. Hence, top 100 means the 100 smallest sum of squared residuals (SSR) between the simulation and observed data
Box plots of top 100 parameters (the same parameter sets as the one used to plot Fig. 5) from the initial cluster (left) and the cluster after 20 iterations of CGN (right). Note the distributions of x5, x8, x9, x10 clearly shrunk after 20 iterations while the distribution of x4 did not change noticeably. (Box plot: The edges of the boxes are the 75th and 25th percentiles. The line in the box is the median, and the whiskers extend to the largest and the smallest value within the 1.5 times the inter-quartile range. Dots are the outliers outside the whiskers)
Scatter plots of parameters. As can be seen in this example, we can find parameter-parameter correlations of some of the parameters found by CGN. (parameters whose corresponding SSR is the least 100 within the cluster of 1,000, i.e., the same parameter sets as the one used to plot the right hand side of Fig. 5)
4 Numerical experiments
In this section we illustrate the advantages of the proposed CGN algorithm by numerical experiments on three PBPK models.
4.1 Numerical experiment setup
In this subsection we specify the details of the numerical experiment set-up.
4.1.1 Mathematical models
For the numerical experiments, we used the following three published mathematical models of drug concentration in the blood of a human body (PBPK model) (Fukuchi et al. 2017; Yao et al. 2018; Yoshikado et al. 2016). The time course drug concentration was simulated using the model, and random noise was added to mimic the observation uncertainties. The random noise was generated by a normal distribution with a standard deviation of 10% of the simulated concentration value. The simulated drug concentration was used as the test dataset, and multiple possible parameters were estimated from the test dataset using CGN and conventional methods. The mathematical description of each PBPK model used for the numerical experiments is summarised in Table 1. Also the detailed description of the model and implementations of all the examples are available as the supplementary material.
4.1.2 Computation environment
All computational experiments were performed using Matlab 2019a on 3.1 GHz Intel Core i5 processors with MacOS version 10.13.6. When using the algorithms implemented in Python we used Python version 3.7. All results of the numerical experiments were summarised and visualised using ggplot2 version 2.2.1 (Wickham 2016) in R version 3.3.2.
4.1.3 The initial set of vectors \(\{\varvec{x}_{i}^{(0)}\}_{i=1}^{N}\)
We generated the initial set of vectors \(\{\varvec{x}_{i}^{(0)}\}_{i=1}^{N}\) randomly based on Algorithm 1. The range of the initial cluster was set by the pharmacologically likely parameter range based on a priori knowledge defined by domain specialists (e.g., from the values obtained from the animal experiments or lab experiments).
We stored and used this initial set of vectors as the initial cluster for the CGN and also as initial iterates for the other nonlinear least squares solvers and optimisation algorithms which were compared.
Notice that this range of the initial cluster does not necessary contain the global minimiser as many pharmacokinetics parameter can be few order of magnitudes different between species. For the sake of comparison with the constrained optimisation algorithms, we constructed Example 2 so that the initial range contains the set of parameters that we used to create the dataset. This ensures that a global minimiser is in the domain where the initial cluster is made. When comparing with the constrained optimisation algorithms, we used this domain as the bound constraint for the parameter search for these algorithms.
4.1.4 ODE solver
For Examples 1 and 3 the nonlinear function evaluations require the numerical solution of stiff systems of ODEs. We used the ODE15s solver ( Shampine and Reichelt 1997) with the default settings to solve these ODEs (relative tolerance \(10^{-3}\), absolute tolerance \(10^{-6}\)). We observed that for some set of parameters, the ODE solver can get stuck in an infinite loop. Here, we set the timeout, where if the ODE evaluation takes longer than 5 seconds, it terminates the ODE evaluation process and returns a not-a-number vector.
4.1.5 Setting for the cluster Gauss–Newton (CGN) method:
We used the following parameters unless stated otherwise:
Here \(k_\text {max}\) is the maximum number of iterations of the method. We used the initial set of vectors \(\{\varvec{x}_{i}^{(0)}\}_{i=1}^{N}\) described in Sect. 4.1.3 as the initial cluster. Note that we chose \(\lambda _\text {init}\) to be consistent with the default setting of the LM implementation (lsqnonlin in Matlab.) In “Appendicies A, B, and C”, we have presented the numerical experiments on the influence of the user-defined algorithm parameters.
4.2 Algorithms compared
Here, we list the conventional and recently developed algorithms that we compared our proposed algorithm with. We used the default setting of the algorithms unless stated otherwise.
4.2.1 Nonlinear least squares solvers
We compare the proposed algorithm against gradient-based and gradient-free nonlinear least squares algorithm s. We used these algorithms to find multiple approximate minimisers of the nonlinear least square problem of our interest by repeatedly applying these algorithms with various initial iterates. Namely; we used each parameter vector in \(\{\varvec{x}_{i}^{(0)}\}_{i=1}^{N}\), which were randomly generated in the CGN method as the initial iterate, and excecuted each algorithm repeatedly to obtain N sets of estimated parameter vectors.
Levenberg–Marquardt (LM) method The conventional gradient-based nonlinear least squares solver, Levenberg–Marquardt method (Björck 1996) implemented in the lsqcurvefit function in Matlab.
We use LM with the default setting as well as setting ‘FiniteDifferenceStepSize’ to be the square root of the default accuracy of the ODE solve (e.g., \(\text {FiniteDifferenceStepSize}=\sqrt{\text {AbsTol}}=\sqrt{10^{-6}}\)). Note that the FiniteDifferenceStepSize of the default setting is \(10^{-6}\).
Trust-Region (TR) method The conventional gradient-based nonlinear least squares solver, Trust-Region method (Conn et al. 2000) implemented in the lsqcurvefit function in Matlab.
DFO–LS method A gradient-free nonlinear least squares solver DFO-LS method (Cartis et al. 2019). We obtained the Python code of DFO-LS Version 1.0.2 from http://people.maths.ox.ac.uk/robertsl/dfols/ (last accessed on June 6th 2019). To make the numerical method for solving the model ODEs exactly the same, we called the Matlab implementation of the nonlinear function through MATLAB Engine API for Python.
Libensemble with POUNDERS We used the libensemble algorithm (Hudson et al. 2019) that was available at the developer branch of https://github.com/Libensemble/libensemble (last accessed April 25th 2019) together with petsc version 3.10.4 available as https://www.mcs.anl.gov/petsc/index.html. We used POUNDERS as the nonlinear least squares solver. As libensemble was implemented in Python, we used the MATLAB Engine API to call the nonlinear function. POUNDERS algorithm utilises bound constraints for the search domain, and we tested this algorithm only for Example 2.
4.2.2 Optimisation algorithms
We compared the proposed method against the optimisation algorithms that are designed to minimise a function which takes a vector quantity as an input and scalar quantity as output. We applied these algorithms to our nonlinear least squares problem by minimising SSR defined in Eq. (3)
Quasi-Newton method We used the Quasi-Newton method implemented in Matlab’s Optimisation toolbox as the fminunc function. This implementation uses the BFGS formula to update the approximate Hessian.
The following algorithms require bound constraints, and they were tested only for Example 2.
Implicit filter method This is another gradient-free optimisation algorithm. The code was downloaded from https://archive.siam.org/books/se23/ (last accessed June 7th 2019). We used the ‘least-squares’ option with a budget of 200.
Surrogate optimisation method We used the Surrogate optimisation algorithm implemented in Matlab’s Global Optimisation Toolbox as the surrogateopt function. We repeated this algorithm 250 times with distinct random-seed to obtain 250 global minimisers of the nonlinear-least squares problem of our interest.
Particle Swarm We used the Particle Swarm optimisation algorithm implemented in Matlab’s Global Optimisation Toolbox as the particleswarm function. We repeated this algorithm 250 times with distinct random seeds to obtain 250 global minimisers of the nonlinear least squares problem of our interest.
Genetic Algorithm We used the Genetic algorithm implemented in Matlab’s Global Optimisation Toolbox as the ga function. As one run of this algorithm required over 100,000 function evaluations for our examples, we did not repeat this algorithm to obtain multiple global minimisers.
DIRECT A sampling algorithm DIRECT (Jones et al. 1993). The Matlab implementation was obtained from https://searchcode.com/codesearch/view/12449743/ (last accessed July 30th 2019). As one run of this algorithm required over 100,000 function evaluations for our examples, we did not repeat this algorithm to obtain multiple global minimisers.
4.3 Results
We compared the proposed CGN method with various existing algorithms. We compared the ‘speed’ of the algorithm by the number of function evaluations required and we compared the ‘quality’ of the minimisers found by the SSR (smaller the SSR the better minimiser). As we can see in Table 2, the dominant part of the computation time was spent by the nonlinear function evaluations in CGN. Hence, we claim that the number of function evaluations is a fair way to compare the computation cost.
We compared the speed for finding acceptable minimisers. We define the acceptable minimisers as the minimisers with SSR less than the SSR of the parameter that was used to generate the test dataset. In Fig. 8, we show the number of acceptable minimisers found given the total number of function evaluations. As can be seen, CGN is significantly faster than any other method for finding acceptable minimisers. In general, the nonlinear least squares solvers were faster than using optimisation algorithms to minimise SSR. Also, some optimisation methods could not find or could only find a few acceptable minimisers. Among the various least squares solvers, the derivative-free methods (CGN, DFO LS, and libensemble POUNDERS) were usually faster than the derivative-based methods.
As this analysis depends on how we define acceptable minimisers, we plotted the number of minimisers found for a given SSR threshold in Fig. 9. For this analysis each algorithm was run until its default stopping criterion (given in the reference of each method) was met. We summarised the number of function evaluations that each algorithm required to meet its default stopping criterion in Table 3.
As can be seen in Fig. 9, for Examples 1 and 3, CGN finds more approximate minimsers with small SSR than all the conventional algorithms. For Example 2, as we know that at least one global minimiser is enclosed in the domain defined by \(\varvec{x}^\text {U}\) and \(\varvec{x}^\text {L}\), we were able to apply optimisation algorithms with bound constraints (libsensemble with POUNDERS, implicit filter, surrogate optimisation, and particle swarm). For this problem, particle swarm obtained slightly more approximate minimisers with small SSR than CGN. However, as can be seen in Table 3, particle swarm required significantly more function evaluations than the CGN (particle swarm: 3,646,900 v.s. CGN: 6768). Aside from the particle swarm, CGN outperformed all the other methods in the number of acceptable minimisers found.
In addition to above rigorous comparison, we applied two global optimisation algorithms, Genetic algorithm (GA) and DIRECT to Example 2. GA and DIRECT required 120,600 and 100,063 nonlinear function evaluations, respectively, to obtain one minimiser each. The SSR of the minimisers found by GA and DIRECTwere 0.0197 and 0.244, respectively, while the minimum SSR found by the CGN is 0.0188. Hence, even to find just one minimiser, CGN is faster and more accurate than these two methods.
Number of acceptable minimisers (out of 250) found by each method for given total number of function evaluations. Acceptable minimisers are defined by the minimisers with SSR less than the SSR of the parameter that was used to generate the test dataset. (Example1: \(\text {SSR}<0.0592\), Example2: \(\text {SSR}<0.0444\), Example3: \(\text {SSR}<0.0915\))
5 Concluding remarks
We proposed the Cluster Gauss–Newton (CGN) method, a new derivative free method specifically designed for finding multiple approximate minimisers of a nonlinear least squares problem. The development of this algorithm was motivated by the parameter estimation of physiologically based pharmacokinetic (PBPK) models that appears in pharmaceutical drug development. The particular nature of the model, where the model is over-parameterised, and consideration of multiple possible parameters is necessary, motivated us to develop the new method. The fact that our algorithm obtains multiple approximate minimisers collectively makes it significantly more computationally efficient compared to existing nonlinear least squares solvers or optimisation algorithms. In addition, we observed that in general, CGN obtains minimisers with smaller sum of squared residuals (SSR) than existing algorithms. We demonstrated these advantages using three examples that come from real world drug development projects.
By minimising the assumption on the nonlinear function, where it can be a “black box”, we have ensured the ease of use and implementation for those who may not have a substantial background in mathematics or scientific computing. We believe this advantage of the proposed method will be appreciated by potential users of the algorithm in industry. In this paper, we used the pharmacokinetics models as examples. However, as we do not assume any particular form of the nonlinear function, we believe the proposed method can be used for many other mathematical models in various scientific fields.
CGN performs sufficiently well for the current use in pharmacokinetic model analyses. However, we believe there are various possible extensions of CGN. First, CGN only makes use of the function evaluations from the one previous iteration. On the other hand, we can consider using the function evaluations from all the previous iterations similarly to the multi-secant method (Eyert 1996; Bierlaire and Crittin 2006; Hicken et al. 2017). CGN and multi-secant methods already share similarities; both methods construct global linear approximations from previously conducted nonlinear function evaluations; this gives an error-tolerant nature and computational efficiency. The error-tolerant nature of the CGN can be seen in “Appendix D”, where the CGN converges even if the nonlinear function is evaluated only up to the first decimal place. Given the similarity, combining these two methods may lead to significant improvement. Second, currently, the CGN does not detect if two or more points in the cluster converge to the same points; hence the computation becomes redundant. We may be able to save computational cost even further by introducing the measure where we consider the point in the cluster is similar enough so that to stop the redundant calculation.
Matlab code is available at https://www.mathworks.com/matlabcentral/fileexchange/68798.
References
Aoki Y, Hayami K, De Sterck H, Konagaya A (2011) Cluster Newton method for sampling multiple solutions of an underdetermined inverse problem: parameter identification for pharmacokinetics. NII Tech Rep 2:1–38
Aoki Y, Hayami K, De Sterck H, Konagaya A (2014) Cluster Newton method for sampling multiple solutions of underdetermined inverse problems: application to a parameter identification problem in pharmacokinetics. SIAM J Sci Comput 36(1):B14–B44. https://doi.org/10.1137/120885462
Asami S, Kiga D, Konagaya A (2017) Constraint-based perturbation analysis with cluster Newton method: a case study of personalized parameter estimations with irinotecan whole-body physiologically based pharmacokinetic model. BMC Syst Biol. https://doi.org/10.1186/s12918-017-0513-2
Bierlaire M, Crittin F (2006) Solving noisy, large-scale fixed-point problems and systems of nonlinear equations. Transp Sci 40(1):44–63
Björck Å (1996) Numerical mthods for least squares roblems. SIAM, Philadelphia. https://doi.org/10.1137/1.9781611971484
Boender CGE, Kan AR, Timmer G, Stougie L (1982) A stochastic method for global optimization. Math Program 22(1):125–140
Broyden CG (1970) The convergence of a class of double-rank minimization algorithms 1. General considerations. IMA J Appl Math 6(1):76–90
Cartis C, Roberts L (2019) A derivative-free Gauss–Newton method. Math Program Comput 11(4):631–674
Cartis C, Fiala J, Marteau B, Roberts L (2019) Improving the flexibility and robustness of model-based derivative-free optimization solvers. ACM Transp Math Softw. 3(32):1–41
Conn AR, Gould NI, Toint PL (2000) Trust Region Methods. SIAM, Philadelphia
Eyert V (1996) A comparative study on methods for convergence acceleration of iterative vector sequences. J Comput Phys 124(2):271–285
Feinberg J, Langtangen HP (2015) Chaospy: an open source tool for designing methods of uncertainty quantification. J Comput Sci 11:46–57
Fletcher R (1970) A new approach to variable metric algorithms. Comput J 13(3):317–322
Fukuchi Y, Toshimoto K, Mori T, Kakimoto K, Tobe Y, Sawada T, Asaumi R, Iwata T, Hashimoto Y, Nunoya KI, Imawaka H, Miyauchi S, Sugiyam Y (2017) Analysis of nonlinear pharmacokinetics of a highly Albumin-bound compound: contribution of Albumin-mediated hepatic uptake mechanism. J Pharm Sci. https://doi.org/10.1016/j.xphs.2017.04.052
Gauss CF (1857) Theory of the motion of the heavenly bodies moving about the Sun in conic sections: A translation of Gauss’s“ Theoria Motus’. With an Appendix. Little, Brown and Company, Boston
Gibaldi M, Perrier D (1982) Drugs and the pharmaceutical sciences. In: Pharmacokinetics, vol. 15, pp. 445–449. Marcel Dekker New York
Goldberg DE, Holland JH (1988) Genetic algorithms and machine learning. Mach Learn 3(2):95–99
Goldfarb D (1970) A family of variable-metric methods derived by variational means. Math Comp 24(109):23–26
Gutmann HM (2001) A radial basis function method for global optimization. J Global Optim 19(3):201–227
Hansen PC (2005) Rank-deficient and discrete Ill-posed problems: numerical aspects of linear nversion, vol 4. SIAM, Philadelphia
Hicken JE, Meng P, Dener A (2017) Error-tolerant multisecant method for nonlinearly constrained optimization. arXiv preprint arXiv:1709.06985
Hudson S, Larson J, Wild SM, Bindel D, Navarro JL (2019) libEnsemble user manual. Tech. Rep. Revision 0.5.1, Argonne National Laboratory. https://buildmedia.readthedocs.org/media/pdf/libensemble/latest/libensemble.pdf
Jones DR, Perttunen CD, Stuckman BE (1993) Lipschitzian optimization without the Lipschitz constant. J Optim Theory Appl 79(1):157–181
Kelley CT (2011) Implicit Filtering, vol. 23 in Software Environments and Tools. SIAM, Philadelphia
Kennedy J, Eberhart R (1995) Particle swarm optimization (PSO). In: Procedings of IEEE international conference on neural networks, Perth, Australia, pp. 1942–1948
Kim SJ, Toshimoto K, Yao Y, Yoshikado T, Sugiyama Y (2017) Quantitative analysis of complex drugdrug interactions between Repaglinide and Cyclosporin A /Gemfibrozil using physiologically based pharmacokinetic models with in vitro transporter/enzyme inhibition data. J Pharm Sci. https://doi.org/10.1016/j.xphs.2017.04.063
Larson J, Menickelly M, Wild SM (2019) Derivative-free optimization methods. Acta Numer 28:287–404
Marquardt DW (1963) An algorithm for least-squares estimation of nonlinear parameters. J Soc Indus Appl Math 11(2):431–441
Mezura-Montes E, Coello CAC (2011) Constraint-handling in nature-inspired numerical optimization: past, present and future. Swarm Evolut Comput 1(4):173–194
Moré JJ (1978) The Levenberg–Marquardt algorithm: implementation and theory. In: Watson GA (ed) Numerical analysis. Lecture notes in mathematics, vol 630. Springer, Berlin, Heidelberg, pp 105–116
Nakamura T, Toshimoto K, Lee W, Imamura CK, Tanigawara Y, Sugiyama Y (2018) Application of PBPK modeling and virtual clinical study approaches to predict the outcomes of CYP2D6 genotype-guided dosing of tamoxifen. CPT Pharm Syst Pharmacol. https://doi.org/10.1002/psp4.12307
Shampine LF, Reichelt MW (1997) The MATLAB ODE suite. SIAM J Sci Comput 41(3):1–22. https://doi.org/10.1137/S1064827594276424
Shanno DF (1970) Conditioning of quasi-Newton methods for function minimization. Math Comput 24(111):647–656
Shanno DF, Kettler PC (1970) Optimal conditioning of quasi-Newton methods. Math Comput 24(111):657–664
Toshimoto K, Tomaru A, Hosokawa M, Sugiyama Y (2017) Virtual clinical studies to examine the probability distribution of the AUC at target tissues using physiologically-based pharmacokinetic modeling: Application to analyses of the effect of genetic polymorphism of enzymes and transporters on Irinotecan Ind. Pharm Res 34(8):1584–1600. https://doi.org/10.1007/s11095-017-2153-z
Watanabe T, Kusuhara H, Maeda K, Shitara Y, Sugiyama Y (2009) Physiologically based pharmacokinetic modeling to predict transporter-mediated clearance and distribution of Pravastatin in humans. J Pharmacol Exp Ther. https://doi.org/10.1124/jpet.108.146647
Wickham H (2016) ggplot2: Elegant graphics for data analysis. Springer-Verlag, New York. http://ggplot2.org
Wild SM (2017) Chapter 40, POUNDERS in TAO: solving derivative-free nonlinear least-squares problems with POUNDERS. Advances and trends in optimization with engineering applications. SIAM, Philadelphia, pp 529–539
Yao Y, Toshimoto K, Kim SJ, Yoshikado T, Sugiyama Y (2018) Quantitative analysis of complex drug-drug interactions between Cerivastatin and metabolism/transport inhibitors using physiologically based pharmacokinetic modeling. Drug Metab Dispos 46(7):924–933
Yoshida K, Maeda K, Kusuhara H, Konagaya A (2013) Estimation of feasible solution space using Cluster Newton Method: application to pharmacokinetic analysis of irinotecan with physiologically-based pharmacokinetic models. BMC Syst Biol 7(Suppl 3):S3. https://doi.org/10.1186/1752-0509-7-S3-S3
Yoshikado T, Yoshida K, Kotani N, Nakada T, Asaumi R, Toshimoto K, Maeda K, Kusuhara H, Sugiyama Y (2016) Quantitative analyses of hepatic OATP-mediated interactions between statins and inhibitors using PBPK modeling with a parameter optimization method. Clin Pharmacol Ther 100(5):513–523
Acknowledgements
We would like to thank Professor Akihiko Konagaya for giving us the opportunity to initiate this research. We would like to thank Dr. Jeffrey Larson for his help on numerical experiment using libensemble. We would like to thank Professor Hiroshi Yabe and Dr. Ryota Kobayashi for useful discussions. In addition, we wish to thank the reviewers of this paper, especially for the ideas for the further improvement of the algorithm. Ken Hayami was supported in part by JSPS KAKENHI Grant Number 15K04768 and 19H03392. Yasunori Aoki is currently employed by AstraZeneca.
Funding
Open access funding provided by Uppsala University.
Author information
Authors and Affiliations
Corresponding author
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Appendices
Appendix A
1.1 Numerical experiments on the influence of the weight of the linear approximation
To illustrate the influence of the weight \(d_{j(i)}^{(k)}\) of the weighted linear approximation in (2-1) of the algorithm [Eqs. (17) and (23)], we conducted numerical experiments using Example 1 of Sect. 4. For this numerical experiment, we varied the parameter \(\gamma \ge 0\) in Eq. (23):
In Fig. 10, we show the number of minimisers found by CGN for given accuracy threshold. As can be seen from the figure , the weight for the linear approximation improves the accuracy and the speed of CGN. Note that \(\gamma =0\), which corresponds to giving equal weights to all the cluster points is not optimal. In this example, \(\gamma \ge 1\) gave good results.
Appendix B
1.1 Numerical experiments on the influence of the regularisation
To illustrate the necessity and the influence of the regularisation in (2-2) of the algorithm [Eq. (26)], we conducted numerical experiments using Example 1 of Sect. 4. We varied the initial value of the regularisation coefficient \(\lambda _\text {init}\) and tested CGN. Figure 11 shows the number of minimisers found by CGN for given accuracy threshold. As can be seen from the figure, regularisation is necessary for CGN to perform well. For this example, \(\lambda _\text {init}=0.1\) gave a good result.
Appendix C
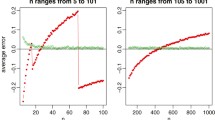
1.1 Numerical experiments on the influence of the initial distribution
To investigate the influence of the distribution of the points in the initial cluster, we have conducted numerical experiments using Example 1 with varying the number of points in the cluster (N = 50, 250, 1000) and type of distribution(Latin hypercube sampling, Sobol sequence, Halton sequence and uniform random sampling). We have used chaospy (Feinberg and Langtangen 2015) and pyDOE (https://pythonhosted.org/pyDOE/) modules on Python version 3.7.1 to generate initial distributions. As can be seen in Fig. 12, when there is a sufficient number of points in the cluster (e.g., N = 1000), then quasi-random sampling technique such as Latin hypercube or Sobol sequence outperformed the uniform random sampling. On the other hand, the uniform random sampling performed consistently well across all numbers of points in the cluster we have tested.
Appendix D
1.1 Application to discontinuous nonlinear functions
The Jacobian based local optimisation methods cannot solve nonlinear least squares problems if the nonlinear function is discontinuous. On the other hand, the Cluster Gauss–Newton (CGN) method uses the linear approximation of the nonlinear function to capture the global behaviour of the function. Hence, it does not require the nonlinear function to be continuous.
For the following numerical experiment, we consider the case where the nonlinear function is not continuous. This example is constructed to show that CGN can solve nonlinear least squares problems that the conventional Jacobian based method cannot solve. We create such a nonlinear function by rounding the nonlinear function of Example 1 [i.e., Eqs. (1)–(3) in the supplementary document] to the first decimal place.
As can be seen in Fig. 13, the CGN method was able to find many accurate solutions. On the other hand, the LM failed to find any reasonable solution. Unlike derivative based LM, a derivative-free method (DFO LS method) occasionally finds reasonable solutions.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Aoki, Y., Hayami, K., Toshimoto, K. et al. Cluster Gauss–Newton method. Optim Eng 23, 169–199 (2022). https://doi.org/10.1007/s11081-020-09571-2
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11081-020-09571-2