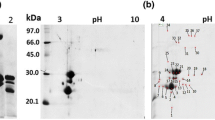
Abstract
Natural rubber or latex from the Hevea brasiliensis is an important commodity in various economic sectors in today’s modern society. Proteins have been detected in latex since the early twentieth century, and they are known to regulate various biological pathways within the H. brasiliensis trees such as the natural rubber biosynthesis, defence against pathogens, wound healing, and stress tolerance. However, the exact mechanisms of the pathways are still not clear. Proteomic analyses on latex have found various proteins and revealed how they fit into the mechanisms of the biological pathways. In the past three decades, there has been rapid latex protein identification due to the improvement of latex protein extraction methods, as well as the emergence of two-dimensional gel electrophoresis (2-DE) and mass spectrometry (MS). In this manuscript, we reviewed the methods of latex protein extraction that keeps on improving over the past three decades as well as the results of numerous latex protein identification and quantitation.


Similar content being viewed by others
Change history
21 March 2023
A Correction to this paper has been published: https://doi.org/10.1007/s10265-023-01451-x
References
Abd-Rahman N, Kamarrudin MF (2018) Proteomic profiling of Hevea latex serum induced by ethephon stimulation. J Trop Plant Physiol 10:11–22
Agrawal AA, Konno K (2009) Latex: a model for understanding mechanisms, ecology, and evolution of plant defense against herbivory. Annu Rev Ecol Evol Syst 40:311–331
Amalou Z, Bangratz J, Chrestin H (1992) Ethrel (ethylene releaser)-induced increases in the adenylate pool and transtonoplast ΔpH within Hevea latex cells. Plant Physiol 98:1270–1276
Bottier C (2020) Biochemical composition of Hevea brasiliensis latex: a focus on the protein, lipid, carbohydrate and mineral contents. In: Robert N (ed) Adv Bot Res, vol 93. Academic Press, London, pp 201–237
Chow KS, Wan KL, Isa MNM, Bahari A, Tan SH, Harikrishna K, Yeang HY (2007) Insights into rubber biosynthesis from transcriptome analysis of Hevea brasiliensis latex. J Exp Bot 58:2429–2440
Chow KS, Mat-Isa MN, Bahari A, Ghazali AK, Alias H, Mohd-Zainuddin Z, Hoh CC, Wan KL (2012) Metabolic routes affecting rubber biosynthesis in Hevea brasiliensis latex. J Exp Bot 63:1863–1871
Chunliu C, Jie Y, Zhi D, Shoucai C, Dejun L (2010) Differential analysis of 2-DE profiles and preliminary identification of lutoids proteins from TPD tree in Hevea brasiliensis. Chin Agric Sci Bull 26:304–308
Cornish K, Backhaus RA (1990) Rubber transferase activity in rubber particles of guayule. Phytochemistry 29:3809–3813
Cornish K, Siler DJ (1996) Characterization of cis-prenyl transferase activity localized in a buoyant fraction of rubber particles from Ficus elastica latex. Plant Physiol Biochem 34:377–384
Cornish K, Wood DF, Windle JJ (1999) Rubber particles from four different species, examined by transmission electron microscopy and electron-paramagnetic-resonance spin labeling, are found to consist of a homogeneous rubber core enclosed by a contiguous, monolayer biomembrane. Planta 210:85–96
Cornish K, Scott DJ, Xie W, Mau CJ, Zheng YF, Liu XH, Prestwich GD (2018) Unusual subunits are directly involved in binding substrates for natural rubber biosynthesis in multiple plant species. Phytochemistry 156:55–72
Coupé M, Lambert C (1977) Absorption of citrate by the lutoids of latex and rubber production by Hevea. Phytochemistry 16:455–458
Dai L, Xiang Q, Li Y, Nie Z, Kang G, Duan C, Zeng R (2012) Rubber particle protein analysis of Hevea brasiliensis by two dimensional 16-BAC/SDS-PAGE and mass spectrometry. Zhongguo Nong Ye Ke Xue 45:2328–2338
Dai L, Kang G, Li Y, Nie Z, Duan C, Zeng R (2013) In-depth proteome analysis of the rubber particle of Hevea brasiliensis (para rubber tree). Plant Mol Biol 82:155–168
Dai L, Kang G, Nie Z, Li Y, Zeng R (2015) Comparative proteomic analysis of latex from Hevea brasiliensis treated with Ethrel and methyl jasmonate using iTRAQ-coupled two-dimensional LC–MS/MS. J Proteom 132:167–175
D’Auzac J, Prévôt JC, Jacob C (1995) What’s new about lutoids? A vacuolar system model from Hevea latex. Plant Physiol Biochem 33:765–777
Dejun L, Jie Y, Zhi D, Chunliu C, Peng H, Min W, Shoucai C (2009) Evaluation of three methods for protein extraction suitable for 2-DE of Hevea brasiliensis C-serum. Chin Agric Sci Bull 25:273–279
Dennis MS, Light DR (1989) Rubber elongation factor from Hevea brasiliensis. Identification, characterization, and role in rubber biosynthesis. J Biol Chem 264:18608–18617
Dian K, Sangare A, Diopoh JK (1995) Evidence for specific variation of protein pattern during tapping panel dryness condition development in Hevea brasiliensis. Plant Sci 105:207–216
Duan CF, Nie ZY, Zeng RZ (2006) Establishment of 2-DE system and primary analyses on the membrane proteins of rubber particles in Hevea brasiliensis by MALDI-TOF. Chin J Tropic Crops 27:22–29
Faurobert M, Pelpoir E, Chaïb J (2007) Phenol extraction of proteins for proteomic studies of recalcitrant plant tissues. In: Zivy M (ed) Plant proteomics, methods and protocols, vol 355. Humana Press, New Jersey, pp 9–14
Gao L, Sun Y, Wu M, Wang D, Wei J, Wu B, Wang G, Wu W, Jin X, Wang X, He P (2018) Physiological and proteomic analyses of molybdenum-and ethylene-responsive mechanisms in rubber latex. Front Plant Sci 9:621
Gemperline E, Keller C, Li L (2016) Mass spectrometry in plant-omics. Anal Chem 88:3422–3434
Gidrol X, Chrestin H, Tan HL, Kush ANIL (1994) Hevein, a lectin-like protein from Hevea brasiliensis (rubber tree) is involved in the coagulation of latex. J Biol Chem 269:9278–9283
Habib MAH, Yuen GC, Othman F, Zainudin NN, Latiff AA, Ismail MN (2017) Proteomics analysis of latex from Hevea brasiliensis (clone RRIM 600). Biochem Cell Biol 95:232–242
Habib MAH, Gan CY, Abdul Latiff A, Ismail MN (2018) Unrestrictive identification of post-translational modifications in Hevea brasiliensis latex. Biochem Cell Biol 96:818–824
Havanapan PO, Bourchookarn A, Ketterman AJ, Krittanai C (2015) Comparative proteome analysis of rubber latex serum from pathogenic fungi tolerant and susceptible rubber tree (Hevea brasiliensis). J Proteomics 131:82–92
Jacob JL, d’Auzac J, Prevot JC (1993) The composition of natural latex from Hevea brasiliensis. Clin Rev Allergy 11:325
Kanokwiroon K, Teanpaisan R, Wititsuwannakul D, Hooper AB, Wititsuwannakul R (2008) Antimicrobial activity of a protein purified from the latex of Hevea brasiliensis on oral microorganisms. Mycoses 51:301–307
Kekwick RGO (2018) The formation of isoprenoids in Hevea latex. In: d’Auzac J (ed) Physiology of rubber tree latex: The laticiferous cell and latex—a model of cytoplasm. CRC Press, Florida, pp 145–164
Kerche-Silva LE, Cavalcante DGSM, Danna CS, Gomes AS, Carrara IM, Cecchini AL, Yoshihara E, Job AE (2017) Free-radical scavenging properties and cytotoxic activity evaluation of latex C-serum from Hevea brasiliensis RRIM 600. Free Radic Antioxid 7:107–114
Ko JH, Chow KS, Han KH (2003) Transcriptome analysis reveals novel features of the molecular events occurring in the laticifers of Hevea brasiliensis (para rubber tree). Plant Mol Biol 53:479–492
Konno K (2011) Plant latex and other exudates as plant defense systems: roles of various defense chemicals and proteins contained therein. Phytochemistry 72:1510–1530
Li HL, Guo D, Lan FY, Tian WM, Peng SQ (2011) Protein differential expression in the latex from Hevea brasiliensis between self-rooting juvenile clones and donor clones. Acta Physiol Plant 33:1853–1859
Liengprayoon S, Vaysee L, Jantarasunthorn S, Wadeesirisak K, Chaiyut J, Srisomboon S, Musigamart N, Roytrakul S, Bonfilis F, Char C, Bottier C (2017) Fractionation of Hevea brasiliensis latex by centrifugation: (i) a comprehensive description of the biochemicalcomposition of the 4 centrifugation fractions. IRRDB 2017. Jakarta 1:645–660
Martin MN (1991) The latex of Hevea brasiliensis contains high levels of both chitinases and chitinases/lysozymes. Plant Physiol 95:469–476
Men X, Wang F, Chen GQ, Zhang HB, Xian M (2019) Biosynthesis of natural rubber: current state and perspectives. Int J Mol Sci 20:50
Oh SK, Kang H, Shin DH, Yang J, Chow KS, Yeang HY, Wagner B, Breiteneder H, Han KH (1999) Isolation, characterization, and functional analysis of a novel cDNA clone encoding a small rubber particle protein from Hevea brasiliensis. J Biol Chem 274:17132–17138
Posch A, Chen Z, Wheeler C, Dunn MJ, Raulf-Heimsoth M, Baur X (1997) Characterization and identification of latex allergens by two-dimensional electrophoresis and protein microsequencing. J Allergy Clin Immunol 99:385–395
Priyadarshan PM (2017) Latex production, diagnosis and harvest. In: Priyadarshan PM (ed) Biology of hevea rubber, 2nd edn. CAB International, Wallingford, pp 51–82
Rahman AYA, Usharraj AO, Misra BB, Thottathil GP, Jayasekaran K, Feng Y, Hou S, Ong SY, Ng FL, Lee LS, Tan HS (2013) Draft genome sequence of the rubber tree Hevea brasiliensis. BMC Genom 14:75
Sansatsadeekul J, Sakdapipanich J, Rojruthai P (2011) Characterization of associated proteins and phospholipids in natural rubber latex. J Biosci Bioeng 111:628–634
Siler DJ, Goodrich-Tanrikulu M, Cornish K, Stafford AE, McKeon TA (1997) Composition of rubber particles of Hevea brasiliensis, Parthenium argentatum, Ficus elastica, and Euphorbia lactiflua indicates unconventional surface structure. Plant Physiol Biochem 35:881–889
Singh AP, Wi SG, Chung GC, Kim YS, Kang H (2003) The micromorphology and protein characterization of rubber particles in Ficus carica, Ficus benghalensis and Hevea brasiliensis. J Exp Bot 54:985–992
Slomkowski S, Alemán JV, Gilbert RG, Hess M, Horie K, Jones RG, Kubisa P, Meisel I, Moemann W, Penczek S, Stepto RF (2011) Terminology of polymers and polymerization processes in dispersed systems (IUPAC Recommendations 2011). Pure Appl Chem 83:2229–2259
Spence D (1908) On the presence of oxydases in india-rubber, with a theory in regard to their function in the latex. Biochem J 3:165
Srisomboon S, Wadeesirisak K, Sauvage FX, Rattanaporn K, Sriroth K, Vaysse L, Bonfils F, Sainte-Beuve J, Liengprayoon S, Bottier C (2014) Optimization of protein extraction from different latex samples of Hevea brasiliensis. Thaksin Univ J 17:26
Subroto T, de Vries H, Schuringa JJ, Soedjanaatmadja UM, Hofsteenge J, Jekel PA, Beintema JJ (2001) Enzymic and structural studies on processed proteins from the vacuolar (lutoid-body) fraction of latex of Hevea brasiliensis. Plant Physiol Biochem 39:1047–1055
Tupý J (1973) The regulation of invertase activity in the latex of Hevea brasiliensis Muell. Arg: the effects of growth regulators, bark wounding, and latex tapping. J Exp Bot 24:516–524
Wang X, Shi M, Lu X, Ma R, Wu C, Guo A, Peng M, Tian W (2010) A method for protein extraction from different subcellular fractions of laticifer latex in Hevea brasiliensis compatible with 2-DE and MS. Proteome Sci 8:35
Wang X, Shi M, Wang D, Chen Y, Cai F, Zhang S, Wang L, Tong Z, Tian WM (2013) Comparative proteomics of primary and secondary lutoids reveals that chitinase and glucanase play a crucial combined role in rubber particle aggregation in Hevea brasiliensis. J Proteome Res 12:5146–5159
Wang X, Wang D, Sun Y, Yang Q, Chang L, Wang L, Meng X, Huang Q, Jin X, Tong Z (2015) Comprehensive proteomics analysis of laticifer latex reveals new insights into ethylene stimulation of natural rubber production. Sci Rep 5:13778
Wang D, Sun Y, Tong Z, Yang Q, Chang L, Meng X, Wang L, Tian W, Wang X (2016) A protein extraction method for low protein concentration solutions compatible with the proteomic analysis of rubber particles. Electrophoresis 37:2930–2939
Wang D, Xie Q, Sun Y, Tong Z, Chang L, Yu L, Zhang X, Yuan B, He P, Jin X, Dong Y (2019) Proteomic landscape has revealed small rubber particles are crucial rubber biosynthetic machines for ethylene-stimulation in natural rubber production. Int J Mol Sci 20:5082
Wiese S, Reidegeld KA, Meyer HE, Warscheid B (2007) Protein labeling by iTRAQ: a new tool for quantitative mass spectrometry in proteome research. Proteomics 7:340–350
Wu X, Xiong E, Wang W, Scali M, Cresti M (2014) Universal sample preparation method integrating trichloroacetic acid/acetone precipitation with phenol extraction for crop proteomic analysis. Nat Prot 9:362
Xiang Q, Xia K, Dai L, Kang G, Li Y, Nie Z, Duan C, Zeng R (2012) Proteome analysis of the large and the small rubber particles of Hevea brasiliensis using 2D-DIGE. Plant Physiol Biochem 60:207–213
Yagami T, Haishima Y, Tsuchiya T, Tomitaka-Yagami A, Kano H, Matsunaga K (2004) Proteomic analysis of putative latex allergens. Int Arch Allergy Immunol 135:3–11
Yamashita S, Yamaguchi H, Waki T, Aoki Y, Mizuno M, Yanbe F, Ishii T, Funaki A, Tozawa Y, Miyagi-Inoue Y, Fushihara K (2016) Identification and reconstitution of the rubber biosynthetic machinery on rubber particles from Hevea brasiliensis. elife 5:e19022
Yan J, Chen SC (2008a) Preliminary analysis of the proteins expressed differentially of lutoid on the latex of tapping panel dryness (TPD) in rubber tree (Hevea brasiliensis Muell. Arg.). North Hortic 7:58–62
Yan J, Chen SC (2008b) Differential analysis of two-dimensional gel electrophoresis profiles and preliminary identification of lutoid proteome on the latex of TPD in Hevea brasiliensis. Hubei Agric Sci 47:858–862
Yan J, Chen SC, Xia ZH (2008) Screening and identification of differential expressed proteins in C-serum on latex of tapping panel dryness (TPD) in rubber tree (Hevea brasiliensis Muell. Arg.). China Biotechnol 28:28–36
Yeang HY, Arif SAM, Yusof F, Sunderasan E (2002) Allergenic proteins of natural rubber latex. Methods 27:32–45
Yuan K, Xu ZJ, Wang ZH, Yang LF (2012) Identification and analysis of latex proteins related to tapping panel dryness in Hevea brasiliensis. J Northwest Univ 27(6):105–109+127
Yuan K, Zhou X, Wang Z, Yang L (2014) Identification and analysis of latex rubber particle proteins related with tapping panel dryness in Hevea brasiliensis. J Nanjing For Univ (Natural Sciences Edition) 38:36–40
Acknowledgements
This work was supported by the Ministry of Higher Education (MOHE) Malaysia under the Fundamental Research Grant Scheme (FRGS) (203.CABR.6711674). We would like to acknowledge USM Fellowship for supporting the PhD studentship of Mohd Afiq Hazlami Habib.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare no conflicts of interest.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Habib, M.A.H., Ismail, M.N. Hevea brasiliensis latex proteomics: a review of analytical methods and the way forward. J Plant Res 134, 43–53 (2021). https://doi.org/10.1007/s10265-020-01231-x
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10265-020-01231-x