Abstract
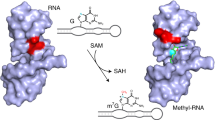
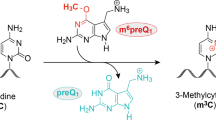
Nearly all classes of coding and non-coding RNA undergo post-transcriptional modification, including RNA methylation. Methylated nucleotides are among the evolutionarily most-conserved features of transfer (t)RNA and ribosomal (r)RNA1,2. Many contemporary methyltransferases use the universal cofactor S-adenosylmethionine (SAM) as a methyl-group donor. SAM and other nucleotide-derived cofactors are considered to be evolutionary leftovers from an RNA world, in which ribozymes may have catalysed essential metabolic reactions beyond self-replication3. Chemically diverse ribozymes seem to have been lost in nature, but may be reconstructed in the laboratory by in vitro selection. Here we report a methyltransferase ribozyme that catalyses the site-specific installation of 1-methyladenosine in a substrate RNA, using O6-methylguanine as a small-molecule cofactor. The ribozyme shows a broad RNA-sequence scope, as exemplified by site-specific adenosine methylation in various RNAs. This finding provides fundamental insights into the catalytic abilities of RNA, serves a synthetic tool to install 1-methyladenosine in RNA and may pave the way to in vitro evolution of other methyltransferase and demethylase ribozymes.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
Data availability
All data generated and analysed during this study are included in this published Article and its Supplementary Information. Source data are provided with this paper.
References
Motorin, Y. & Helm, M. RNA nucleotide methylation. Wiley Interdiscip. Rev. RNA 2, 611–631 (2011).
Waddell, T. G., Eilders, L. L., Patel, B. P. & Sims, M. Prebiotic methylation and the evolution of methyl transfer reactions in living cells. Orig. Life Evol. Biosph. 30, 539–548 (2000).
Jadhav, V. R. & Yarus, M. Coenzymes as coribozymes. Biochimie 84, 877–888 (2002).
Frye, M., Jaffrey, S. R., Pan, T., Rechavi, G. & Suzuki, T. RNA modifications: what have we learned and where are we headed? Nat. Rev. Genet. 17, 365–372 (2016).
Traube, F. R. & Carell, T. The chemistries and consequences of DNA and RNA methylation and demethylation. RNA Biol. 14, 1099–1107 (2017).
Zaccara, S., Ries, R. J. & Jaffrey, S. R. Reading, writing and erasing mRNA methylation. Nat. Rev. Mol. Cell Biol. 20, 608–624 (2019).
Frye, M., Harada, B. T., Behm, M. & He, C. RNA modifications modulate gene expression during development. Science 361, 1346–1349 (2018).
Bohnsack, K. E., Höbartner, C. & Bohnsack, M. T. Eukaryotic 5-methylcytosine (m5C) RNA methyltransferases: mechanisms, cellular functions, and links to disease. Genes (Basel) 10, 102 (2019).
Becker, S., Schneider, C., Crisp, A. & Carell, T. Non-canonical nucleosides and chemistry of the emergence of life. Nat. Commun. 9, 5174 (2018).
Schneider, C. et al. Noncanonical RNA nucleosides as molecular fossils of an early Earth-generation by prebiotic methylations and carbamoylations. Angew. Chem. Int. Ed. 57, 5943–5946 (2018).
Higgs, P. G. & Lehman, N. The RNA world: molecular cooperation at the origins of life. Nat. Rev. Genet. 16, 7–17 (2015).
Doudna, J. A. & Cech, T. R. The chemical repertoire of natural ribozymes. Nature 418, 222–228 (2002).
Pyle, A. M. Group II intron self-splicing. Annu. Rev. Biophys. 45, 183–205 (2016).
Ren, A., Micura, R. & Patel, D. J. Structure-based mechanistic insights into catalysis by small self-cleaving ribozymes. Curr. Opin. Chem. Biol. 41, 71–83 (2017).
Attwater, J., Raguram, A., Morgunov, A. S., Gianni, E. & Holliger, P. Ribozyme-catalysed RNA synthesis using triplet building blocks. eLife 7, e35255 (2018).
Wachowius, F., Attwater, J. & Holliger, P. Nucleic acids: function and potential for abiogenesis. Q. Rev. Biophys. 50, e4 (2017).
Tjhung, K. F., Shokhirev, M. N., Horning, D. P. & Joyce, G. F. An RNA polymerase ribozyme that synthesizes its own ancestor. Proc. Natl Acad. Sci. USA 117, 2906–2913 (2020).
Wilson, C. & Szostak, J. W. In vitro evolution of a self-alkylating ribozyme. Nature 374, 777–782 (1995).
Sharma, A. K. et al. Fluorescent RNA labeling using self-alkylating ribozymes. ACS Chem. Biol. 9, 1680–1684 (2014).
Ameta, S. & Jäschke, A. An RNA catalyst that reacts with a mechanistic inhibitor of serine proteases. Chem. Sci. (Camb.) 4, 957–964 (2013).
McDonald, R. I. et al. Electrophilic activity-based RNA probes reveal a self-alkylating RNA for RNA labeling. Nat. Chem. Biol. 10, 1049–1054 (2014).
Peselis, A. & Serganov, A. Themes and variations in riboswitch structure and function. Biochim. Biophys. Acta 1839, 908–918 (2014).
McCown, P. J., Corbino, K. A., Stav, S., Sherlock, M. E. & Breaker, R. R. Riboswitch diversity and distribution. RNA 23, 995–1011 (2017).
Breaker, R. R. Riboswitches and translation control. Cold Spring Harb. Perspect. Biol. 10, a032797 (2018).
Batey, R. T. Recognition of S-adenosylmethionine by riboswitches. Wiley Interdiscip. Rev. RNA 2, 299–311 (2011).
Sun, A. et al. SAM-VI riboswitch structure and signature for ligand discrimination. Nat. Commun. 10, 5728 (2019).
Burke, D. H. & Gold, L. RNA aptamers to the adenosine moiety of S-adenosyl methionine: structural inferences from variations on a theme and the reproducibility of SELEX. Nucleic Acids Res. 25, 2020–2024 (1997).
Lindahl, T., Demple, B. & Robins, P. Suicide inactivation of the E. coli O 6-methylguanine-DNA methyltransferase. EMBO J. 1, 1359–1363 (1982).
Ghaem Maghami, M., Scheitl, C. P. M. & Höbartner, C. Direct in vitro selection of trans-acting ribozymes for posttranscriptional, site-specific, and covalent fluorescent labeling of RNA. J. Am. Chem. Soc. 141, 19546–19549 (2019).
Ghaem Maghami, M., Dey, S., Lenz, A. K. & Höbartner, C. Repurposing antiviral drugs for orthogonal RNA-catalyzed labeling of RNA. Angew. Chem. Int. Ed. 59, 9335–9339 (2020).
Keppler, A. et al. A general method for the covalent labeling of fusion proteins with small molecules in vivo. Nat. Biotechnol. 21, 86–89 (2003).
Dalhoff, C., Lukinavicius, G., Klimasăuskas, S. & Weinhold, E. Direct transfer of extended groups from synthetic cofactors by DNA methyltransferases. Nat. Chem. Biol. 2, 31–32 (2006).
Oerum, S., Dégut, C., Barraud, P. & Tisné, C. m1A post-transcriptional modification in tRNAs. Biomolecules 7, 20 (2017).
Chujo, T. & Suzuki, T. Trmt61B is a methyltransferase responsible for 1-methyladenosine at position 58 of human mitochondrial tRNAs. RNA 18, 2269–2276 (2012).
Reichle, V. F., Weber, V. & Kellner, S. NAIL-MS in E. coli determines the source and fate of methylation in tRNA. ChemBioChem 19, 2575–2583 (2018).
Filonov, G. S., Kam, C. W., Song, W. & Jaffrey, S. R. In-gel imaging of RNA processing using broccoli reveals optimal aptamer expression strategies. Chem. Biol. 22, 649–660 (2015).
Xiong, X., Li, X. & Yi, C. N 1-methyladenosine methylome in messenger RNA and non-coding RNA. Curr. Opin. Chem. Biol. 45, 179–186 (2018).
Grozhik, A. V. et al. Antibody cross-reactivity accounts for widespread appearance of m1A in 5′UTRs. Nat. Commun. 10, 5126 (2019).
Safra, M. et al. The m1A landscape on cytosolic and mitochondrial mRNA at single-base resolution. Nature 551, 251–255 (2017).
Zhou, H. et al. Evolution of a reverse transcriptase to map N 1-methyladenosine in human messenger RNA. Nat. Methods 16, 1281–1288 (2019).
Terns, M. P. & Terns, R. M. Small nucleolar RNAs: versatile trans-acting molecules of ancient evolutionary origin. Gene Expr. 10, 17–39 (2002).
Serganov, A. et al. Structural basis for discriminative regulation of gene expression by adenine- and guanine-sensing mRNAs. Chem. Biol. 11, 1729–1741 (2004).
Gilbert, S. D., Reyes, F. E., Edwards, A. L. & Batey, R. T. Adaptive ligand binding by the purine riboswitch in the recognition of guanine and adenine analogs. Structure 17, 857–868 (2009).
Pitsch, S., Weiss, P. A., Jenny, L., Stutz, A. & Wu, X. Reliable chemical synthesis of oligoribonucleotides (RNA) with 2′-O-[(triisopropylsilyl)oxy]methyl(2′-O-tom)-protected phosphoramidites. Helv. Chim. Acta 84, 3773–3795 (2001).
Wachowius, F. & Höbartner, C. Probing essential nucleobase functional groups in aptamers and deoxyribozymes by nucleotide analogue interference mapping of DNA. J. Am. Chem. Soc. 133, 14888–14891 (2011).
Košutić, M. et al. A mini-twister variant and impact of residues/cations on the phosphodiester cleavage of this ribozyme class. Angew. Chem. Int. Ed. 54, 15128–15133 (2015).
Höbartner, C. et al. The synthesis of 2′-O-[(triisopropylsilyl)oxy] methyl (TOM) phosphoramidites of methylated ribonucleosides (m1G, m2G, m2 2G, m1I, m3U, m4C, m6A, m6 2A) for use in automated RNA solid-phase synthesis. Monatshefte Chemie 134, 851–873 (2003).
Rio, D. C. Expression and purification of active recombinant T7 RNA polymerase from E. coli. Cold Spring Harb. Protoc. 2013, pdb.prot078527 (2013).
Mortimer, S. A. & Weeks, K. M. A fast-acting reagent for accurate analysis of RNA secondary and tertiary structure by SHAPE chemistry. J. Am. Chem. Soc. 129, 4144–4145 (2007).
Fujii, T., Itaya, T. & Saito, T. Purines. 18. Kinetic studies of base-catalysed conversion of 1-alkyladenosines into N-alkyladenosines – effect of substituents on rearrangement rate. Chem. Pharm. Bull. (Tokyo) 23, 54–61 (1975).
Acknowledgements
This work was supported by the European Research Council (ERC-CoG 682586) and by the Deutsche Forschungsgemeinschaft (DFG) (SPP1784 Chemical Biology of native nucleic acid modifications). We thank J. Adelmann and S. Mayer for help with mass spectrometric analyses, C. Steinmetzger for synthesis of 1M7 and S. Dey for providing m6dG.
Author information
Authors and Affiliations
Contributions
In vitro selection was carried out by C.P.M.S. RNA solid-phase synthesis was performed by A.-K.L. and C.H. Ribozymes were characterized by C.P.M.S., M.G.M. and A.-K.L. Plasmids were constructed by M.G.M., A.-K.L. and C.P.M.S. RNA structure probing and detection of RNA methylation by primer extension was performed by C.P.M.S. LC–MS analyses were run by C.P.M.S. and C.H. C.P.M.S., M.G.M. and C.H. designed experiments. C.P.M.S. and C.H. wrote the paper. All authors analysed data and commented on the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Peer review information Nature thanks Pål Falnes and the other, anonymous, reviewer(s) for their contribution to the peer review of this work. Peer reviewer reports are available.
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data figures and tables
Extended Data Fig. 1 In vitro selection of methyltransferase ribozymes.
a, In vitro selection scheme consisting of incubation, capture, wash, elution, amplification and transcription steps. The RNA substrate (blue) contains an unpaired adenosine (red, A) and is connected to the RNA library via the single-stranded loop (black). The library contains 40 random nucleotides (green) and 2 constant binding arms (cyan) complementary to the RNA substrate upstream and downstream of the bulged A. Incubation was 50 μM RNA, 100 μM BG–biotin, 50 mM HEPES, pH 7.5, 120 mM KCl, 5 mM NaCl and 40 mM MgCl2, at 37 °C (round 1–8: 16 h; round 9–11: 4 h; and round 11: 50 μM BG–biotin). For capture, beads were blocked with E. coli tRNA; streptavidin and neutravidin beads were switched every other round. Denaturing wash buffer was 8 M urea, 10 mM Tris-HCl, pH 7.5, 1 mM EDTA, 0.01% Tween-20. Elution used 95% formamide, 1 mM EDTA, at 95 °C for 5 min. b, For RT–PCR, the following conditions were used: 42 °C, 30 min, 10 cycles of PCR with 1 μM primer D4 and 0.5 μM primer D5. For the following PCR, we used: 25 cycles, 5% (v/v) RT–PCR product as template, 1 μM D4 and 0.5 μM D3, 10% (v/v) DMSO, and an annealing temperature of 65 °C. For in vitro transcription, we used a dsDNA template from a 200 μl PCR reaction, 100 μl reaction volume with 4 mM each NTP, followed by PAGE purification.
Extended Data Fig. 2 Activity of methyltransferase ribozymes.
Examination of mutations in the stem-loop. a, 3′-Fluorescein-labelled R10a tested with 100 μM m6G. b, 5′-Fluorescein-labelled R1 tested with 100 μM BG-NH2. ON, overnight (23 h). c, Kinetics of CA13 (Rz1/Rz1s)-catalysed alkylation of R1 using BG-NH2 cofactor. Fraction modified is shown as mean ± s.d. (n = 3), and fit to a mono-exponential model (Y = Ymax(1 – e–kt)) (blue) or a bi-exponential model (Y = Ymax(a (1 – e–k1t) + (1 – a) (1 – e–k2t)) (black). d, Dependence of MTR1 methylation yield on Mg2+ concentration; reactions were performed with 100 μM m6G (on R2 with Rz3) at 37 °C. Individual data points are shown as empty symbols (n = 2 for 5 h and 23 h, n = 3 for 7 h time points), and mean is depicted as a filled symbol.
Extended Data Fig. 3 RNA structure probing by DMS and SHAPE.
MTR1 (Rz3) was annealed with 17-nt RNA (R6), treated with DMS or 1M7, in presence (+) or absence (–) of m6G, and the modification pattern was analysed by primer extension (5′-32P-labelled D4) with Superscript III. DMS probes the accessibility of the Watson–Crick face of A and C, and SHAPE with 1M7 probes the flexibility of the backbone. Both probing methods confirm the central base-paired stem and reveal the protection of several additional nucleotides (bold). The experiment was repeated three times with similar results.
Extended Data Fig. 4 Mutagenesis of target RNA.
Representative gel images of RNA-catalysed alkylation reactions of RNA-substrate mutants by their corresponding ribozymes, with complementary binding arms as listed in Supplementary Table 1. a, Adenosine point mutations. b, Atomic mutagenesis of backbone. c, d, Atomic mutagenesis of adenosine. e, Reaction sites blocked by methylation. f, Binding-arm mutations outside of GAG. g, Point mutations next to the target nucleoside A. Reactions were performed with 100 μM BG-NH2 (a–e) or BG–biotin (f, g) at pH 7.5, 40 mM MgCl2 and 37 °C, and repeated two times for each substrate. The parent reaction with adenosine was performed with fluorescently labelled and radioactively labelled RNA independently six times.
Extended Data Fig. 5 RNA-catalysed methylation of tRNAs.
a, tRNA sequences studied. b, Synthetic fragments and corresponding ribozymes. c, Exemplary gel images for kinetic analysis of MTR1-catalysed fragment methylation showing quantitative formation of m1A. d, Exemplary HR-ESI-MS of isolated methylated R. norvegicus RNA fragment. e, Full gel images of primer extension analyses shown in Fig. 3. Representative gel images of three independent experiments with similar results.
Extended Data Fig. 6 Specificity of ribozyme for the target tRNA.
a, Secondary structure schemes of six E. coli tRNAs, with very similar TΨC-stem-loop sequences (drawn without natural modifications). Target tRNAAsp in top left corner, with A58 indicated in blue. The nucleotides complementary to the binding arms of MTR1–tRNAAsp(A58) (Rz23) are shown in bold. The purple nucleotides indicate mismatched positions with the binding arms. b, Full gel image of primer extension analysis on total E. coli tRNA with six primers, shown in Fig. 3c. Primer extension analyses were repeated twice.
Extended Data Fig. 7 Plasmid-encoded cis-active ribozyme.
a, Schematic of FBC–MTR1 construct. b, Dot plot for FBC–MTR1 generated by Vienna RNAfold (http://rna.tbi.univie.ac.at/), indicating a high probability of folding into the designed structure. c, LC–MS analysis of the in vitro transcript FBC–MTR1 digested by snake venom phosphodiesterase and bacterial alkaline phosphatase, after reaction with m6G. EIC for detection of MH+ 282.11 ± 0.05 (methylated adenosines) shows production of m1A, and m6A to a small extent (due to partial Dimroth rearrangement during digestion). The bottom trace for the synthetic references m1A and m6A (50 nM each) is the same as shown in Fig. 3d. d, Primer extension stop assays also confirm activity of FBC–MTR1 transcribed in vitro and in vivo, in the presence of total E. coli RNA. Left, full gel image shown in Fig. 4a for in vitro-transcribed FBC–MTR1. Right, primer extension on total E. coli RNA, isolated 1 h after IPTG induction and incubated with the indicated m6G or BG concentration in vitro. These experiments were independently repeated two times with similar results.
Extended Data Fig. 8 Plasmid-encoded trans-active ribozyme.
a, Schematic of FBT–MTR1 with specific binding arms for E. coli tRNAAsp (A58). b, Primer extension stop assays confirm the activity and specificity of the FBT–MTR1 in vitro transcript. Left, full gel image shown in Fig. 4b for FBT–MTR1 reacted with m6G and BG on total E. coli tRNA. Right, primer extension on BG-treated sample with six different E. coli tRNA-specific primers (P4–P9), repeated twice. c, LC–MS analysis after digestion of total E. coli tRNA treated with FBT–MTR1 and BG (same sample as for the right gel image). EIC for MH+ 282.11 (methylated adenosines) and 298.11 (methylated guanosines) demonstrate the presence of natural tRNA modifications. EIC 358.11 in comparison to reference nucleosides bn1A and bn6A shows bn-modified adenosines produced by FBT–MTR1.
Supplementary information
Supplementary Figure 7
-page pdf file showing uncropped images (full scans) of gels used in Figures 1b-d, 2e, and ED Fig 2a-c, 4a-g, 5c. The regions used are indicated by red boxes and labeled with the used RNA substrate and ribozyme, according to the numbers in Supplementary Table 1.
Supplementary Table
This file contains Supplementary Tables 1-3.
Rights and permissions
About this article
Cite this article
Scheitl, C.P.M., Ghaem Maghami, M., Lenz, AK. et al. Site-specific RNA methylation by a methyltransferase ribozyme. Nature 587, 663–667 (2020). https://doi.org/10.1038/s41586-020-2854-z
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41586-020-2854-z
This article is cited by
-
Programmable RNA base editing via targeted modifications
Nature Chemical Biology (2024)
-
Ribozyme for stabilized SAM analogue modifies RNA in cells
Nature Chemistry (2023)
-
A SAM analogue-utilizing ribozyme for site-specific RNA alkylation in living cells
Nature Chemistry (2023)
-
A new RNA performs old chemistry
Nature Chemical Biology (2022)
-
Structural basis for substrate binding and catalysis by a self-alkylating ribozyme
Nature Chemical Biology (2022)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.