The Synergistic Effect of Mud Crab Antimicrobial Peptides Sphistin and Sph12−38 With Antibiotics Azithromycin and Rifampicin Enhances Bactericidal Activity Against Pseudomonas Aeruginosa
- 1State Key Laboratory of Marine Environmental Science, College of Ocean & Earth Sciences, Xiamen University, Xiamen, China
- 2State-Province Joint Engineering Laboratory of Marine Bioproducts and Technology, College of Ocean & Earth Sciences, Xiamen University, Xiamen, China
Overuse or abuse of antibiotics has undoubtedly accelerated the increasing prevalence of global antibiotic resistance crisis, and thus, people have been trying to explore approaches to decrease dosage of antibiotics or find new antibacterial agents for many years. Antimicrobial peptides (AMPs) are the ideal candidates that could kill pathogens and multidrug-resistant bacteria either alone or in combination with conventional antibiotics. In the study, the antimicrobial efficacy of mud crab Scylla paramamosain AMPs Sphistin and Sph12−38 in combination with eight selected antibiotics was evaluated using a clinical pathogen, Pseudomonas aeruginosa. It was interesting to note that the in vitro combination of rifampicin and azithromycin with Sphistin and Sph12−38 showed significant synergistic activity against P. aeruginosa. Moreover, an in vivo study was carried out using a mouse model challenged with P. aeruginosa, and the result showed that the combination of Sph12−38 with either rifampicin or azithromycin could significantly promote the healing of wounds and had the healing time shortened to 4–5 days compared with 7–8 days in control. The underlying mechanism might be due to the binding of Sphistin and Sph12−38 with P. aeruginosa lipopolysaccharides (LPS) and subsequent promotion of the intracellular uptake of rifampicin and azithromycin. Taken together, the significant synergistic antibacterial effect on P. aeruginosa in vitro and in vivo conferred by the combination of low dose of Sphistin and Sph12−38 with low dose of rifampicin and azithromycin would be beneficial for the control of antibiotic resistance and effective treatment of P. aeruginosa-infected diseases in the future.
Introduction
Pseudomonas aeruginosa is an opportunistic Gram-negative bacterial pathogen and can cause infections and mass mortality in patients that have cystic fibrosis, AIDS, severe burns, organ transplants, and cancer (Lyczak et al., 2000; Blonder et al., 2004). The current treatment regimen of P. aeruginosa includes a wide range of antibiotics including β-lactams, aminoglycosides, fluoroquinolones, or even the inter-combination of those antibiotics (Hancock and Speert, 2000); however, the clinical pathogen P. aeruginosa is less susceptible to almost all of the routinely used antibiotics and fairly easy to develop resistance. For example, from 2003 to 2011, the rates of carbapenem-resistant P. aeruginosa (CRPA) isolated from patients with hospital-acquired infections in a tertiary care hospital in northeast China were 14.3, 17.1, 21.1, 24.6, 37.0, 48.8, 56.4, 51.2, and 54.1% over time (Xu et al., 2013). In another hospital, First Affiliated Hospital of Nanjing Medical University, in 2008, the resistant rates of P. aeruginosa to cephalosporins (Ceftazidime, Cefotaxime, and Cefepime) were 5.9, 82.4, and 17.6%, respectively, while by the end of 2011, only 4 years passed, those numbers increased to 37.8, 85.7, and 27.8%, respectively (Zhang et al., 2015). Owing to its high intrinsic resistance to antibiotics and wide repertoire of virulence factors, the therapy for the P. aeruginosa-infected diseases becomes an intractable issue (Hancock and Speert, 2000). As reported, the resistance of P. aeruginosa is mainly due to the low permeability of its outer membrane (Hancock, 1998). Besides, the transmembrane efflux pumps are also considered for the intrinsic resistance of P. aeruginosa by which the incoming antibiotics can be taken out of the bacteria efficiently (Li et al., 1994). Therefore, exploration of new antipseudomonal agents is desperately needed to take control of the ubiquitous and acute drug resistance of P. aeruginosa.
To date, multifarious highlighted new strategies against the multidrug-resistant (MDR) bacteria have been proposed and some potential biological products or pharmaceuticals are expected to be applied in clinic, including antimicrobial peptides (AMPs), anti-virulence compounds, phage therapy, and new molecules (Pacios et al., 2020). For example, both Enterobacter cloacae (Mu208) and Klebsiella pneumoniae (Mu1343) display multiple heteroresistance to the antibiotics; the simultaneous combinations of antibiotics targeting multiple heteroresistance are effective to kill these two kinds of bacteria, whereas those targeting homogeneous resistance are ineffective (Band et al., 2019). The sequential therapy is considered as a sustainable strategy to counter the antibiotic crisis because this therapeutic method can constrain the emergence of drug resistance and enhance the bactericidal activity (Roemhild and Schulenburg, 2019). Collateral sensitivity means that the mutations in bacteria cause multidrug resistance but simultaneously enhance sensitivity to many other unrelated drugs, and this new mechanism might be developed to alternative antimicrobial strategies against the multidrug bacteria (Pal et al., 2015). AMPs are widespread distributed in various organisms whose many tissues and cell types could produce different functional AMPs (Vizioli and Salzet, 2002; Zasloff, 2002; Brogden et al., 2003). Most AMPs attach to and permeate the target membrane bilayers to induce pore formation and cause the leakage of cytoplasm (Shai, 2002; Brogden, 2005). Besides that, some peptides can alter the septum formation of cytoplasmic membrane and inhibit the synthesis of cell wall, nucleic acid, and protein or enzymatic activity to kill the bacteria (Brogden, 2005). In addition, AMPs can damage the bacterial cell wall, resulting in the radical change of the bacterial morphology, while simple mutations of the bacteria could not reserve the situations (Shai, 2002; Zasloff, 2002; Chongsiriwatana et al., 2008). Although the AMPs possess better antibacterial activity and a broad antibacterial spectrum, antibiotics have not been successfully substituted by AMPs yet. One reason was that the bacteria also developed resistance to AMPs (Habets et al., 2012; Dobson et al., 2013; Makarova et al., 2018; El Shazely et al., 2020); for example, the point mutations induced conformational changes in BraS or BraR, resulting in the constitutive expression of VraDE, conferring Staphylococcus aureus to evolve high resistance to nisin A (Arii et al., 2019). The second reason is that there is also cross-resistance of evolved strains to other AMPs, not much but it still exists; for instance, the melittin-resistant S. aureus displays cross-resistance against pexiganan (El Shazely et al., 2020). However, despite resistance evolution to AMPs conferred by a few bacteria or cross-resistance of evolved strains to other AMPs, according to the pharmacodynamic studies of AMPs, compared with antibiotics, the evolution of resistance to AMPs is much lower (Yu et al., 2018). Therefore, AMPs are considered to be the potential ideal substituents for antibiotics to be used to some extent in the future. Some studies also have shown that the combination of AMPs with conventional antibiotics has synergistic effect against the targeted pathogenic microorganisms (Li et al., 2017; Zheng et al., 2017; Koppen et al., 2019).
Rifampicin is one kind of derivative of rifamycin. It displays a broad spectrum of antibacterial spectrum against Gram-positive bacteria, particularly Mycobacteria and, to a lesser extent, Gram-negative bacteria such as Escherichia coli, Neisseria meningitides, etc (Walter and Staehelin, 1971; Heinz Floss and Yu, 2005). The antibacterial mechanism of rifampicin roots in its high affinity binding to and inhibition of the bacterial DNA-dependent RNA polymerase (Campbell et al., 2001). Azithromycin is a kind of macrolide antibiotic, which has a 15-member macrocyclic lactone ring. It is derived from the erythromycin 14-member ring that is inserted into an amino group (Alvarez-Elcoro and Enzler, 1999). Azithromycin also has a broad spectrum of antibacterial spectrum against Gram-positive bacteria including S. aureus, parts of Streptococci, Streptococcus pneumoniae etc.; Gram-negative bacteria including Haemophilus influenzae, Haemophilus ducreyi, Neisseria gonorrhoeae, Bordetella pertussis, etc.; and other pathogens such as Chlamydia trachomatis, Ureaplasma urealyticum, Mycoplasma pneumoniae, etc (Retsema et al., 1987; Peters et al., 1992; Alvarez-Elcoro and Enzler, 1999). Azithromycin inhibits the bacterial growth by interfering with their protein synthesis. It could also inhibit RNA-dependent protein synthesis by reversibly binding to the 50 S subunits of the bacterial ribosome (Mazzei et al., 1993; Alvarez-Elcoro and Enzler, 1999).
Our previous studies demonstrate that the AMPs Sphistin (Chen et al., 2015) and Sph12−38 (Ma et al., 2017) from the mud crab Scylla paramamosain show potent activity against the hospital-acquired opportunistic pathogen P. aeruginosa (24 and 12 μmol·L−1, respectively). Sphistin is a 38-amino-acid peptide that is derived from the N-terminal of histone H2A in S. paramamosain, and Sph12−38 is a truncated short fragment of Sphistin. This study aimed to understand whether the clinical medicine azithromycin and rifampicin in combination with Sphistin and Sph12−38 would have a synergistic effect on P. aeruginosa. In vitro experiments were performed using Sphistin in combination with each of two selected antibiotics azithromycin and rifampicin. Furthermore, an in vivo study was carried out using a mouse model with wound as infection model and the subsequent treatment was evaluated using Sph12−38 in combination with each of azithromycin and rifampicin.
Materials and Methods
Peptides, Antibiotics, and Bacterial Strains
The peptides Sphistin (AGGKAGKDSGKSKAKAVSRSARAGLQFPVGRIHRHLK; molecular mass, 3828.48 Da) and Sph12−38 (KAKAKAVSRSARAGLQFPVGRIHRHLK; molecular mass, 2983.59 Da) were all synthesized by Shanghai Glory Chemistry Co., Ltd., and the purity of these two peptides reached 98.88% and 98.68%, respectively. The eight medical injections were all purchased from Zhongshan Hospital Xiamen University. The bacterial strain P. aeruginosa (ATCC 9027) was purchased from the CGMCC. Bacterial strains were cultivated in Nutrient broth (NB) overnight at 37°C.
Antimicrobial Activity
After the bacteria were all in logarithmic phase, aliquots of the bacterial cell suspension (~5 × 105 CFU·ml−1) were then added to 96-well plates; each well-contained 100 μl of cell suspension. The peptides and antibiotics were all dissolved in sterile water, and the final concentration of the antibacterial agents ranged from 1.5 to 48 μmol·L−1, and then twofold serial dilutions of the peptide and antibiotics were mixed with the bacteria with an equal volume. The samples were subsequently incubated at 37°C for 24 h. The minimal inhibitory concentrations (MICs) were defined as the lowest concentration of antibacterial agents that completely inhibited bacterial growth. The MICs of the antibacterial agents against the tested microorganisms were determined by the standard broth microdilution method (Khara et al., 2014; Yamamoto and Tamura, 2014).
Synergistic Effect Assay
The synergistic effects of Sphistin and Sph12−38 in combination with the antibiotics were tested using the checkerboard assay as previous research described (Rand et al., 1993; Petersen et al., 2006). Twofold serial dilutions of Sphistin, Sph12−38, and the antibiotics were prepared, the peptides were mixed in a 1:1 volume ratio with the antibiotics, and then the mixture (100 μl) was added into 96-well plates. The equal volume of bacterial suspension (~5 × 105 CFU·ml−1) was seeded into the plates and incubated with the antibacterial agent mixture at 37°C for 24 h. To ensure the precision of experimental results, each assay was in triplicate and repeated three times. The fractional inhibitory concentration index (FICI) was used to assess the synergistic effects of the combination of AMPs with antibiotics. The FICI could be calculated by the formula: FICI = [MICAMPs in synergistic system]/[MICAMPs alone] + [MICAntibiotics in synergistic system]/[MICAntibiotics alone] (Pankey and Ashcraft, 2005; Pankey et al., 2005). When FICI <0.5, it was interpreted as synergy; 0.5 ≤ FICI < 1.0, partial synergy; 1.0 ≤ FICI < 4.0, additive effect; and FICI ≥ 4.0, antagonism (Odds, 2003).
The Time-Course Killing Kinetics
The time-course killing kinetics were assayed using P. aeruginosa (ATCC 9027) in the presence of Sphistin (6 μmol·L−1 1/4 × MIC), azithromycin (18 μg·ml−1 1/10 × MIC), and a combination of Sphistin (6 μmol·L−1 1/4 × MIC) with azithromycin (18 μg·ml−1 1/10 × MIC); for Sphistin and/or rifampicin, they are as follows: Sphistin (1.5 μmol·L−1 1/16 × MIC), rifampicin (0.625 μg·ml−1 1/4 × MIC), and a combination of Sphistin (1.5 μmol·L−1 1/16 × MIC) with rifampicin (0.625 μg·ml−1 1/4 × MIC). The bacterial cells were cultured overnight and further cultured in new medium, the next day to reach the logarithmic phase, and then incubated with Sphistin and/or rifampicin for an additional 0, 5, 15, 30, 45, 60, and 120 min at 37°C; meanwhile, the experimental bacteria were also incubated with Sphistin and/or azithromycin for an additional 0, 30, 60, 90, 120, 240, and 360 min at 37°C. The total treated bacterial population was then plated on NB agar plates and continued to be incubated overnight at 37°C, and finally we counted the colonies.
Live/Dead Assay
The P. aeruginosa strain (ATCC 9027) was cultured at 37°C until the bacterial cells reached the logarithmic phase, and then the bacterial cells were harvested and washed twice using the NB. The pellet was resuspended to ~106 CFU·ml−1 in the same buffer, after which the prepared bacteria were treated as mentioned above. After the bacteria were treated with all the antibacterial agents, all the bacteria were harvested and stained with SYTO 9 and propidium iodide (PI) in the ratio of 1:1 from the LIVE/DEAD® BacLightTM Bacterial Viability Kits (Thermo Fisher Scientific). After mixing all the mixture thoroughly, the CytoFLEX flow cytometry (Beckman Coulter, California, USA) was used to test the cell membrane integrity and cell viability of the bacteria.
Scanning Electron Microscopy
P. aeruginosa (ATCC 9027) cells in mid-log phase were suspended in PBS to ~1 × 107 CFU·ml−1, after which aliquots were treated as mentioned above. The control group was treated with DPBS (2.45 g Na2HPO4·12H2O and 0.49 g NaH2PO4·2H2O dissolved in 1,000 ml of sterile water, pH = 7.4). After incubation, the bacterial cell pellets were harvested and fixed in 2.5% glutaraldehyde for 2 h at 4°C, followed by two washes in DPBS. The fixed cells were dehydrated for 15 min using a graded ethanol series (30, 50, 70, 90, and 100%). Then, the cells were dehydrated for 5 min in tertiary butanol, this operation was repeated 10 times, and finally, the samples were immersed in tertiary butanol overnight at 4°C. When the prepared specimens dried, conductive coating was applied to the specimens and they were examined using a field emission scanning electron microscopy (SUPRA 55; ZEISS, Germany).
Bacterial Cell Membrane Permeabilization Assay
The permeability of bacterial cell membranes was determined by measuring the leakage of intracellular ATP levels out of the bacterial cells as described by previous research (Koshlukova et al., 1999). Briefly, P. aeruginosa (ATCC 9027) was cultured overnight at 37°C, the cells were harvested and washed twice, and the bacterial cells were resuspended in DPBS. The prepared bacterial cells were treated as mentioned above. After incubation, samples were centrifuged to get the supernatant and then 10 μl of the supernatant was added into 90 μl of the standard reaction solution that comes from the Molecular Probes' ATP Determination Kit (Thermo Fisher Scientific). Prior to testing the luminescence of the samples, use the luminometer to measure the background luminescence and then subtract the background luminescence and read the luminescence of the samples. Using the gradient dilution ATP standard solution to generate a standard curve for a series of ATP concentrations, and according to the standard curve, we could calculate the amounts of the leakage of the intracellular ATP.
Transmission Electron Microscopy (TEM)
The treated bacteria were fixed in 2.5% glutaraldehyde overnight at 4°C, the samples were washed in PBS, the bacteria were harvested and resuspended in PBS (1.5 × 109 CFU·ml−1), and the samples were added into the agar models. The mixture was centrifuged and the supernatant was removed, and the prepared samples were put into 2% molten agar solution, after which the mixed soulution was served on ice until agar solution solidification. The agar block was cut into the size of a rice grain and washed twice, and the agar granules were resuspended and fixed in 2.5% glutaraldehyde overnight at 4°C. Finally, the fixed agar granules were suspended in PBS; after embedding and sectioning, the samples were examined by TEM (Tecnai G2 Spirit, FEI, USA).
Closure of Wounds Infected With P. aeruginosa
Six to eight week old BALB/c male mice weighting 25–28 g (n = 42) were used in the study. The wound was produced by using the medical pressure-sensitive adhesive tape to remove a 2 cm × 2 cm area of the epidermis on the backs of mice. P. aeruginosa (ATCC 9027) (1 × 108 CFU per 20 μl in PBS) was then immediately smeared onto the artificial wound. Two hours after bacterial infection at the wound site, rifampicin (1.25 μg·ml−1, 1/2 × MIC), azithromycin (90 μg·ml−1, 1/2 × MIC), Sph12−38 (24 μmol·L−1, 2 × MIC) alone and in combination with rifampicin (1.25 μg·ml−1, 1/2 × MIC), and azithromycin (90 μg·ml−1, 1/2 × MIC), respectively, in 20 μl of PBS were administered in the wound site by hypodermic injection. The mice without infection of P. aeruginosa (ATCC 9027) were used as uninfected controls. The wounds were photographed at a definite time to record the wound healing.
Statistical Analysis
All experiments were performed three independent times, with each sample performed in triplicate. All data were expressed as means ± standard deviations. Differences among groups were evaluated by using one-way analysis of variance. P < 0.05 were considered statistically significant.
Results
The Synergistic and Additive Antibacterial Effects of Sphistin and Sph12−38 in Combination With Eight Commonly Used Antibiotics
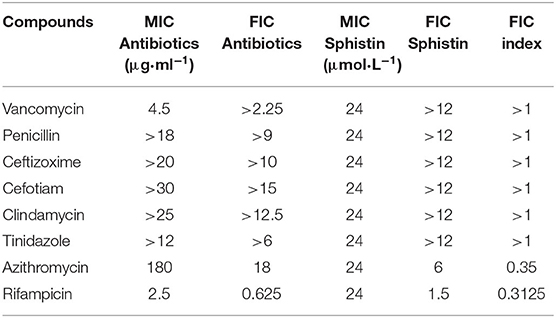
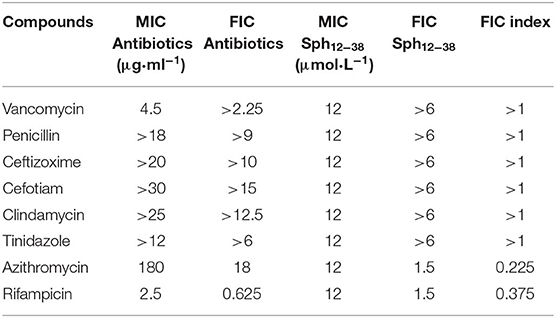
As reported previously (Chen et al., 2015), the synthetic Sphistin has no cytotoxicity toward mouse osteoblastic cell MC3T3-E1 and crab hemocytes even at high tested concentrations (100 mg·ml−1). Similarly, Sph12−38 also exhibits no cytotoxicity on HeLa cell and crab hemocytes (Ma et al., 2017). Both of the AMPs had strong antibacterial activity and also showed potent activity against P. aeruginosa, whose MIC values were 24 and 12 μmol·L−1, respectively. The antimicrobial activities of Sphistin and Sph12−38 in combination with the commonly used clinical antibiotics rifampicin, vancomycin, penicillin, ceftizoxime, cefotiam, clindamycin, tinidazole, and azithromycin against P. aeruginosa were individually determined using the broth microdilution method in accordance with the Clinical and Laboratory Standards Institute (CLSI) recommendation (C. L. S. Institute, 2012), and the results are summarized in Tables 1, 2. Among the eight selected antibiotics, only Sphistin and Sph12−38 in combination with azithromycin and rifampicin exhibited significant synergistic activity against P. aeruginosa. The mixture of 4-fold reduction of Sphistin (reduced from 24 to 6 μmol·L−1) with 10-fold reduction of azithromycin (reduced from 180 to 18 μg·ml−1) and the mixture of 16-fold reduction of Sphistin (reduced from 24 to 1.5 μmol·L−1) with 4-fold reduction of rifampicin (reduced from 2.5 to 0.625 μg·ml−1) could inhibit the growth of P. aeruginosa. Similar situations also occurred when Sph12−38 in combination with azithromycin and rifampicin and the mixture of 8-fold reduction of Sph12−38 (reduced from 24 to 1.5 μmol·L−1) with 10-fold reduction of azithromycin (reduced from 180 to 18 μg·ml−1) or with 4-fold reduction of rifampicin (reduced from 2.5 to 0.625 μg·ml−1) could also significantly minimize the growth of P. aeruginosa. The FICIs of these four combinations were all <0.5 (Tables 1, 2), which could be considered as a synergistic effect. However, for other antibiotics, including vancomycin, penicillin, ceftizoxime, cefotiam, clindamycin, and tinidazole, when they were combined with either Sphistin or Sph12−38 to treat P. aeruginosa, the FICIs were all more than 1, which showed no synergistic effect.
Effects of Sphistin With Azithromycin and Rifampicin on Viability of P. aeruginosa
The effects of Sphistin in combination with azithromycin and rifampicin on viability and membrane integrity of P. aeruginosa were tested by using the LIVE/DEAD® BacLightTM Bacterial Viability Kits and flow cytometry. This kit has two-color fluorescence: the SYTO 9 green-fluorescent nucleic acid stain, which could stain all living cells green, and the red-fluorescent nucleic acid stain, PI, which could specifically penetrate the bacterial cells such that the cell membrane is damaged and cells are stained red. When the flow cytometry was used to detect the mixture of the same number of living bacteria and completely dead bacteria, the living bacteria were stained with SYTO 9, and they were all almost distributed in the Q1-LR quadrant, while the completely dead bacteria were stained with PI, and they were all almost distributed in the Q1-UL quadrant (Figure 1G). The results showed that ~46.01% and ~7.36% of the bacteria treated with Sphistin and azithromycin, respectively, were stained by PI (Figures 1A,B), and the combination of Sphistin and azithromycin could totally kill 85.93% of the bacterial cells (Figure 1C). As for Sphistin and/or rifampicin treatment, only ~7.73% and ~20.06% of the bacterial cells were completely killed by Sphistin and rifampicin, respectively (Figures 1D,E), while when Sphistin is in combination with rifampicin, ~35.19% of the bacterial cells were completely killed (Figure 1F). These findings indicated that exposure to both Sphistin in combination with azithromycin or rifampicin resulted in the uptake of PI by more bacterial cells than Sphistin and these two antibiotics alone, suggesting a significant increase in cell permeability and hence the synergistic activity of Sphistin in combination with these two antibiotics, especially with the azithromycin.
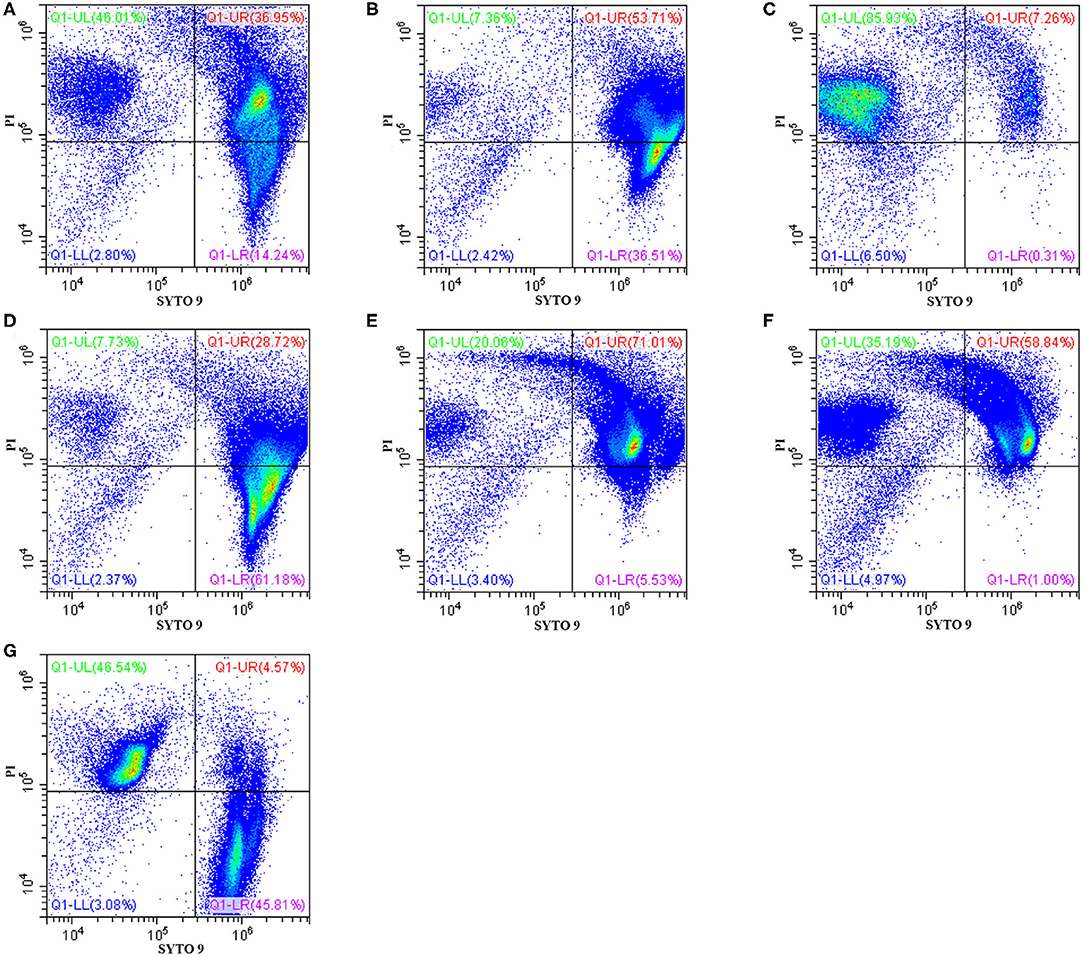
Figure 1. Flow cytometry showing P. aeruginosa exposed to Sphistin, azithromycin, rifampicin, Sphistin in combination with azithromycin, and Sphistin in combination with rifampicin. The bacteria were all incubated with (A) 6 μmol L−1 Sphistin, (B) 18 μg·ml−1 azithromycin, (C) a combination of 6 μmol·L−1 Sphistin and 18 μg·ml−1 azithromycin for 4 h at 37°C; and (D) 1.5 μmol·L−1 Sphistin, (E) 0.625 μg·ml−1 rifampicin, (F) a combination of 1.5 μmol·L−1 Sphistin and 0.625 μg·ml−1 rifampicin for 2 h at 37°C. Then, the bacteria were all stained with SYTO 9 and propidium iodide (PI) fluorescent nucleic acid stain. (G) The same amounts of living bacteria and completely dead bacteria were stained with SYTO 9 and PI, respectively. Bacteria were stained with SYTO 9, which showed living cells in the Q1-LR quadrant, while those bacterial cells that were only stained with PI were all in the Q1-UL quadrant; in the Q1-UR quadrant, the bacterial cells were stained by the SYTO 9 and PI together. The results were detected by flow cytometry.
The Time-Course Killing Kinetics
According to the results, Sphistin in combination with azithromycin and rifampicin had synergistic effects against P. aeruginosa. We also conducted a time-course killing experiment to examine the effects of Sphistin and/or these two antibiotics against P. aeruginosa. Sphistin in combination with azithromycin reduced the number of bacteria by more than two orders of magnitude after 45 min, and after 2 h, all the bacteria were killed. By contrast, the Sphistin or azithromycin used alone did not inhibit the bacterial viability efficiently (Figure 2A). Similarly, Sphistin in combination with rifampicin also inhibited the growth of P. aeruginosa; after 1 h, the number of bacteria was also reduced more than two orders of magnitude, and the combination of Sphistin and rifampicin could kill all the bacteria after 4 h. However, if Sphistin or rifampicin was incubated with the bacteria alone, each of them could not inhibit bacterial viability, and the concentration of bacteria increased after 4 h (Figure 2B).
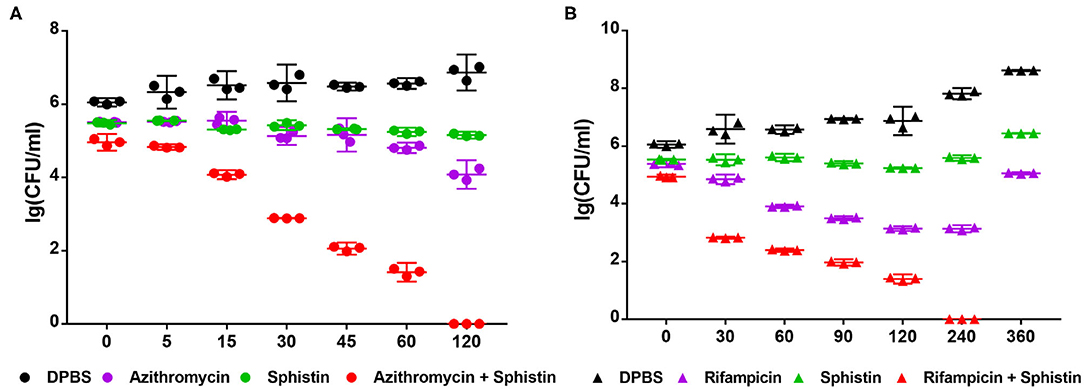
Figure 2. Growth curves of P. aeruginosa when incubated with (A) DPBS, Sphistin (6 μmol·L−1), azithromycin (18 μg·ml−1), and Sphistin (6 μmol·L−1) in combination with azithromycin (18 μg·ml−1) for 2 h; (B) DPBS, Sphistin (1.5 μmol·L−1), rifampicin (0.625 μg·ml−1), and Sphistin (1.5 μmol·L−1) in combination with rifampicin (0.625 μg·ml−1) for 6 h.
Visualization of the Interaction of Sphistin and/or Rifampicin and Azithromycin With P. aeruginosa
Scanning electron microscopy (SEM) was used to visualize the bacterial cell membrane damaged by Sphistin and/or rifampicin and azithromycin. Compared with the control group (Figure 3G), P. aeruginosa treated with Sphistin or azithromycin alone showed slight cell shrinkage (Figures 3A,B), but the cell membrane was intact. When the bacteria were incubated with a combination of Sphistin and azithromycin, the entire cell membrane was completely damaged along with the leakage of cytoplasmic contents (Figure 3C). When Sphistin or rifampicin was used alone, each reagent only induced slight changes in cellular morphology (Figures 3D,E); however, when the bacteria were treated with Sphistin in combination with rifampicin, obvious depressions were observed on the bacterial cell membrane, but no leakage of cytoplasmic content was present and the cellular morphology remained intact (Figure 3F).
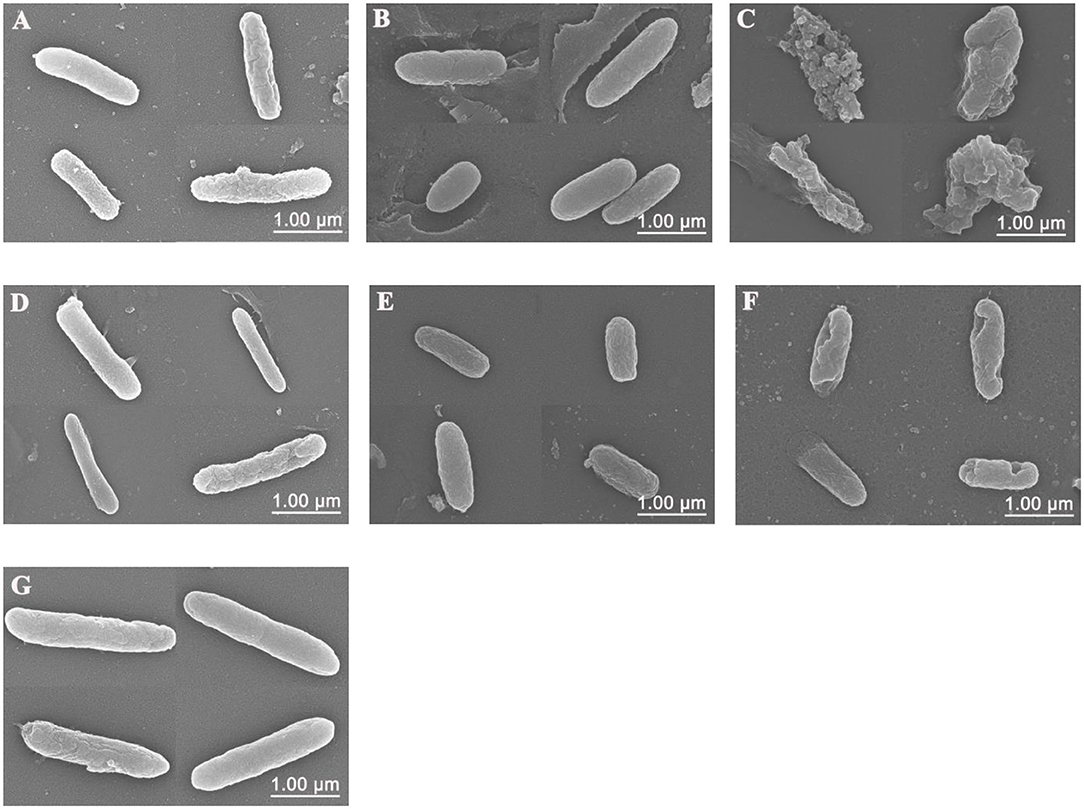
Figure 3. Scanning electron microscope (SEM) images of P. aeruginosa treated with (A) 6 μmol·L−1 Sphistin, (B) 18 μg·ml−1 azithromycin, and (C) a combination of 6 μmol·L−1 Sphistin and 18 μg·ml−1 azithromycin for 2 h at 37°C. Meanwhile, the bacteria treated with (D) 1.5 μmol·L−1 Sphistin, (E) 0.625 μg·ml−1 rifampicin, (F) a combination of 1.5 μmol·L−1 Sphistin and 0.625 μg·ml−1 rifampicin, and (G) DPBS for 4 h at 37°C.
Antimicrobial Mechanism of Sphistin in Combination With Rifampicin and Azithromycin
As reported, when the cell membrane was compromised, the barrier function of the cell membrane will be impaired, resulting in the leakage of critical cellular contents (Khara et al., 2015). To further investigate the mechanism of the combination of Sphistin with rifampicin and azithromycin, we evaluated the changes in membrane permeability by measuring the extracellular ATP levels after the two combination group treatments. Compared with the DPBS group, extracellular ATP could not be detected when both the antibiotics were incubated with P. aeruginosa. However, bacteria treated with Sphistin alone or in combination with the two tested antibiotics can induce the release of ATP from bacterial cells (Figures 4A,B). Moreover, the leakage of intracellular ATP would not increase, regardless of Sphistin in combination with the antibiotics or used alone. Meanwhile, the leakage of intracellular ATP levels would increase with the extension of time, even at lower concentration of Sphistin. In addition, we also used TEM to observe changes in morphology of P. aeruginosa cells or cell membranes. After treatment with DPBS, the control cells had a complete cell morphology (Figure 5A), and no significant changes were observed when the bacterial cells were incubated with Sphistin or azithromycin (Figures 5B,C). The bacteria were then treated with a combination of Sphistin with azithromycin. After 1 h, we observed a significant separation of the cell membrane and cytoplasm in the bacterial cells (Figure 5D, black arrowheads), and remarkable leakage of cellular contents also appeared at the same time (Figure 5D, red arrowheads).
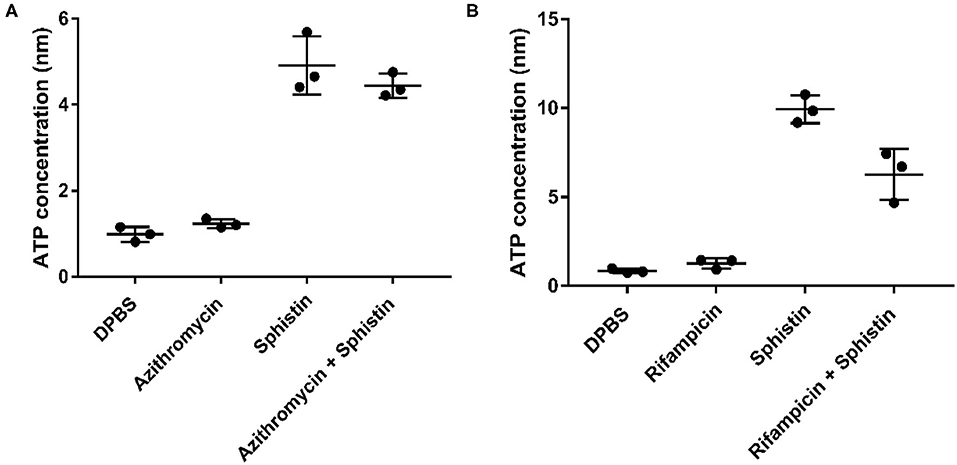
Figure 4. Extracellular ATP release in different treatment groups after exposure of P. aeruginosa to (A) DPBS, Sphistin (6 μmol·L−1), and/or azithromycin (18 μg·ml−1) for 2 h; (B) DPBS, Sphistin (1.5 μmol·L−1), and/or rifampicin (0.625 μg·ml−1) for 4 h. The bacteria cell membrane damage induced by the antibacterial agents is accompanied by leakage of intracellular content due to compromised membrane integrity.
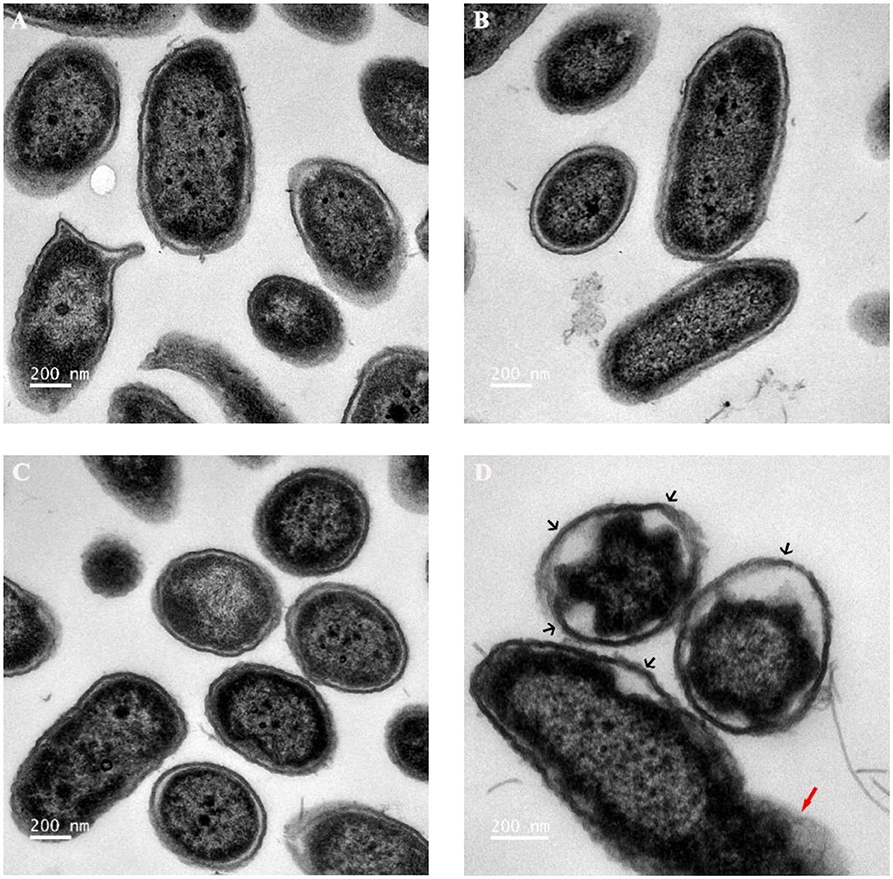
Figure 5. Transmission electron microscopy (TEM) images of P. aeruginosa treated with (A) DPBS, (B) 6 μmol·L−1 Sphistin, (C) 18 μg·ml−1 azithromycin, and (D) a combination of 6 μmol·L−1 Sphistin and 18 μg·ml−1 azithromycin for 1 h at 37°C.
Sph12−38 in Combination With Either Rifampicin or Azithromycin to Promote the Wound Healing
A mouse wound model was used to test the antibacterial activity of Sph12−38 in combination with rifampicin and azithromycin in vivo. On the dorsal part of each mouse, the epidermis was damaged with medical pressure-sensitive adhesive tape and then the dermis was exposed. Afterwards, 108 CFU of the P. aeruginosa were evenly smeared onto the wound areas (Figure 6A). When the wound areas were not infected with P. aeruginosa, only the phosphate-buffered saline (PBS) was injected, and all wounds healed within 5–7 days (Figures 6A,B). Meanwhile, for mice infected with P. aeruginosa, PBS, Sph12−38, rifampicin, azithromycin, and Sph12−38 in combination with rifampicin or azithromycin were injected into the wound skin, respectively. When the infected mice were only inoculated with PBS, the wound healing process became slower and was not completed until 7–8 days postinjection. Compared with the PBS group, the injection of Sph12−38 or rifampicin alone could not significantly shorten the healing time. In contrast, when the infected mice were injected with Sph12−38 in combination with rifampicin, the wound could be completely recovered within 5 to 7 days (P < 0.05). As for azithromycin, when used alone or in combination with Sph12−38, its efficacy seemed to be even better than the combination of Sph12−38 and rifampicin, and the wound completely healed within only 4–5 days (Figures 6A,B).
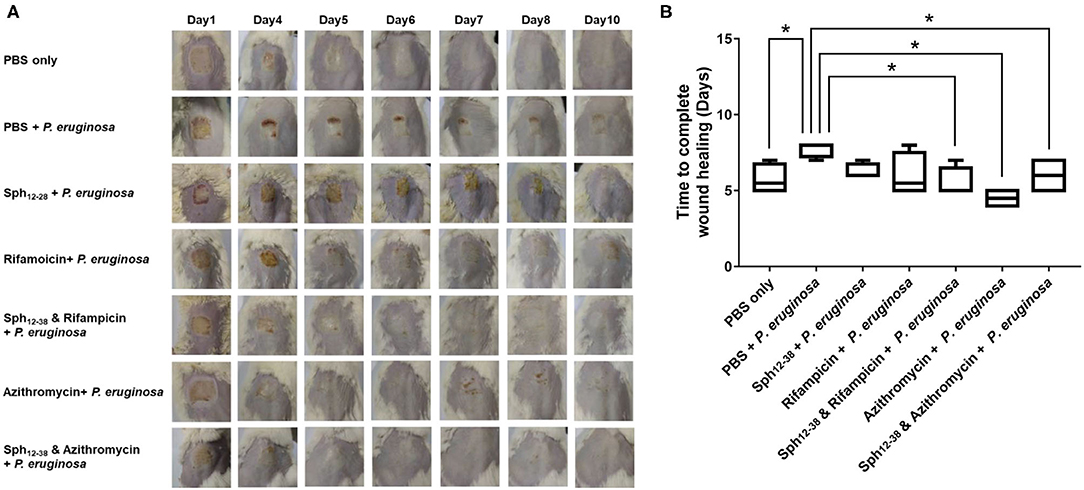
Figure 6. Effect of Sph12−38 in combination with rifampin and azithromycin, respectively, on wound healing in vivo. (A) Insertion status of the wound areas on days 1, 4, 5, 6, 7, 8, and 10 after injury. (B) Time to complete wound healing in each group. Data are presented as the means ± standard deviations (n = 6 mice per group). *P < 0.05 for Sph12−38 in combination with rifampicin-treated mice, Sph12−38 alone, and in combination with azithromycin-treated mice, respectively, vs. PBS-treated mice. Differences among groups were evaluated by using Bonferroni correction of variance.
Discussion
Since penicillin was introduced into clinical treatment in the 1940s, antibiotics abuse has never ended throughout the past 70 years. The abuse of antibiotics, including overuse and misuse of antibiotics, and the limited availability of new antibiotics have caused the global antibiotic resistance crisis (Ventola, 2015). The abuse of antibiotic inevitably leads to the rapid emergence of drug-resistant bacteria including multidrug resistant (MDR) bacteria, and even the extremely drug-resistant (XDR) or totally drug-resistant (TDR) phenotypes worldwide, which have seriously endangered the efficacy of antibiotics and have become a serious threat to human health (French G. L., 2010; Magiorakos et al., 2012). Although it is known that the resistance determinants are already presented in the microorganisms prior to the introduction of antibiotics and most of them are found in natural antibiotic-producing microorganisms (Levy, 1992), the intensive use of antibiotics indeed has dramatically increased the frequency of resistance among nearly all pathogens that greatly weaken therapeutic options and the medical advantages in the post-antibiotic era that had almost been lost to date (Guay, 2008; Lew et al., 2008; Woodford and Livermore, 2009). Therefore, reducing the use of antibiotics had been proposed, through which the selection pressure for acquired resistance will be reduced and the antibiotic-sensitive bacteria will be recovered, enabling them to eventually defeat resistance strains over time (Levin et al., 1997; Andersson and Levin, 1999). AMPs with a membrane targeting effect provide possibilities to use AMPs in combination with multiple antibiotics for treatment of pathogens, thereby enhancing the efficacy of those antibiotics (Cassone and Otvos, 2010; Haney et al., 2017). Among drug-resistant bacteria, P. aeruginosa is one of the most common hospital-acquired and nosocomial conditioned pathogens. It is much prone to acquire multidrug resistance; e.g., some strains of MDR P. aeruginosa have been found to be resistant to almost all antibiotics, including aminoglycosides, cephalosporins, fluoroquinolones, and carbapenems (C. D. C. P. (US) Centres for Disease Control and Prevention (US)., 2013; Frieden, 2013). Therefore, it is necessary to explore new agents that could be substituted for antibiotics.
Rifampicin is a semisynthetic antibiotic derived from rifamycin, and it was introduced as an effective medicine to treat tuberculosis; the primary efficacy of rifampicin was against Gram-positive bacteria (Bliziotis et al., 2007). Several studies have revealed that rifampicin in combination with colistin/polymyxins in vitro (Giamarellos-Bourboulis et al., 2003; Tascini et al., 2004; Yang et al., 2016) and in vivo (Cirioni et al., 2007; Cai et al., 2018) showed effective antibacterial activity against MDR P. aeruginosa. For example, four patients infected with sepsis or pneumonia caused by MDR P. aeruginosa were successfully cured with the addition of rifampicin to colistin (Tascini et al., 2004). The colistin/polymyxins have an amphipathic structure with clusters of hydrophobic and positively charged regions, and this structural property seems to be closely related to their antibacterial activity (Wade et al., 1990; Scott et al., 1999). In fact, the amphipathic structure with hydrophobic and positive charge is also the classical structural feature of the cationic AMPs; therefore, the combination of rifampicin and two α-helical cationic AMPs, magainin II and cecropin A, also showed synergistic antibacterial effect against P. aeruginosa strains in vitro and in vivo (Cirioni et al., 2008). In addition to rifampicin, it has been proven that the combination of colistin and azithromycin showed synergistic and additive activity against the Gram-negative bacteria Acinetobacter baumannii and P. aeruginosa, respectively (Timurkaynak et al., 2006). Two AMPs, Sphistin with 38 aa that is derived from the N-terminal of histone H2A in S. paramamosain and the truncated short fragment Sph12−38, both have potent in vitro antibacterial activity against several Gram-positive and Gram-negative bacteria and some fungi. Both Sphistin and Sph12−38 showed typical features of cationic AMPs, including amphiphilic α-helical second structure and positive charge net. Therefore, both of these two cationic AMPs were used in combination with rifampicin and azithromycin to treat P. aeruginosa in this study.
Outer membranes of Gram-negative bacteria are formed by a divalent cation-crosslinked matrix of lipopolysaccharide (LPS) molecules on the outer leaflet, and via displacing the LPS-bound metals, they could be disrupted by a diverse structural class of polycations (Vaara, 1993; Livermore, 2002; Schuldiner, 2006; Tenover, 2006). It has been proven that polymyxins could bind the lipid A moiety of LPS and perturb the bacterial cell membranes (Evans et al., 1999). Similar to the polymyxins, the cationic AMPs mainly targeted the bacterial cell membranes, and the common features shared by these peptides are that they are prone to form amphipathic structures and then cluster the basic and hydrophobic amino acids into specific regions (Vaara and Porro, 1996; Yeaman and Yount, 2003). Therefore, the cationic AMPs could bind the LPS by electrostatic and hydrophobic interactions and have antibacterial activity (Iwagaki et al., 2000; Yethon and Whitfield, 2001). Meanwhile, the antimicrobial mechanism of Sphistin was suggested to have the capability of attaching the cell membrane and permeabilizing the bacterial cell membranes to kill the pathogens (Chen et al., 2015). Therefore, when P. aeruginosa was treated with the combination of Sphistin and two antibiotics, the permeabilization of the bacterial cell membranes facilitated the uptake of the two antibiotics; rifampicin would bind to and inhibit the bacterial DNA-dependent RNA polymerase (Campbell et al., 2001), and azithromycin would inhibit RNA-dependent protein synthesis of the bacteria (Mazzei et al., 1993; Alvarez-Elcoro and Enzler, 1999); finally, both antibiotics would induce cell death. Just as Figure 4 showed, there was no difference in the leakage of the intracellular ATP between Sphistin used alone or the combination of Sphistin with two antibiotics. Indeed, these results further verified that even in the synergy system, only Sphistin induced the permeabilization of the bacterial cell membranes. Taken together, our results agreed with previous studies (Cirioni et al., 2008) that when the AMPs are combined with the antibiotics to treat the pathogens, the AMPs induced the permeabilization of the pathogen cell membranes and subsequently promoted uptake of the antibiotics, allowing the antibiotics to interact with their intracellular targets easily and finally kill the pathogens.
Among the eight selected antibiotics, the two antibiotics azithromycin and rifampicin in combination with the AMPs Sphistin or Sph12−38 showed a synergistic effect against P. aeruginosa. However, the other six antibiotics had no significant synergistic effect against P. aeruginosa when each of them was combined with the AMPs. Among the six antibiotics, as previously reported (Menninger and Otto, 1982; Nord and Kager, 1983; Gardner and Hill, 2001; Raether and Hanel, 2003; Tasca et al., 2003), tinidazole and clindamycin also have intracellular targets. Among the remaining four antibiotics, the three antibiotics including penicillin, ceftizoxime, and cefotiam are all β-lactam antibiotics, while vancomycin is a glycopeptide antibiotic (Kahne et al., 2005). All of these four antibiotics can inhibit the synthesis of bacteria cell walls as previously reported (Wise and Park, 1965; Waxman et al., 1980; Kahne et al., 2005).
Tinidazole is a structural analog of metronidazole. Both tinidazole and metronidazole are active against anaerobic organisms or protozoa. The antibacterial mechanism is briefly described as follows. When tinidazole diffused into bacterial cells, the nitro group of tinidazole will be reduced to short-lived and toxic free radicals. The toxic intermediates covalently bind to DNA, causing DNA damage and ultimately cell death (Nord and Kager, 1983; Gardner and Hill, 2001; Raether and Hanel, 2003; Tasca et al., 2003). With the decrease of intracellular concentration of tinidazole due to the reduction reaction, more tinidazole could enter the cells, thereby maintaining the inhibition activity of anaerobic bacteria. In aerobic bacteria and mammalian cells, they have relatively high redox potentials and are also rich in oxygen molecules than anaerobic organisms or protozoa, which will hinder the reduction reaction (Nord and Kager, 1983; Tasca et al., 2003). Because P. aeruginosa is an aerobic bacterium, we speculated that tinidazole will not have much effect on P. aeruginosa due to the lack of anaerobic condition in cells. Therefore, even if tinidazole is used in combination with either Sphistin or Sph12−38, more tinidazole might get access into the bacterial cells, but tinidazole cannot or rarely show effective bactericidal activity against P. aeruginosa. Meanwhile, the concentration of the AMPs in the synergistic system was only 1/2 MIC, which cannot inhibit P. aeruginosa alone as testified in the study. Generally, only when the concentration of the AMPs was equal to or greater than the MIC could the AMPs effectively inhibit the target bacteria. Thus, although when in combination with the concentration of 1/2 MIC, Sphistin or Sph12−38 can also accelerate entrance of tinidazole into P. aeruginosa, it still did not produce a significant synergistic effect.
Another antibiotic with intracellular action, clindamycin, belongs to lincosamide, which is a 50S ribosome inhibitor. It inhibits bacteria by preventing peptidyl-tRNAs from entering the ribosome and finally triggers the dissociation of the peptidyl-tRNA (Menninger and Otto, 1982). As reported in previous studies, in general, aerobic Gram-negative (G–) bacteria are resistant to clindamycin, but clindamycin is effective against the Gram-positive (G+) bacteria, such as S. aureus, Streptococcus pyogenes, S. pneumoniae, and Streptococcus viridans. The synergistic effect of clindamycin combined with AMPs has been reported previously (Spizek et al., 2004; Nguschwemlein et al., 2014; Chernysh et al., 2018). For the G+ bacteria, when clindamycin was used in combination with the AMPs cyclooctapeptides (CPs, including CPs 1–3, 5–7, 10, 11) against S. aureus, all combinations showed partial synergistic effects (0.5 ≤ FICI < 1) (Nguschwemlein et al., 2014). When clindamycin was used in combination with FLIP7, the AMP complex from the blowfly Calliphora vicina contains a combination of defensins, cecropins, diptericins, and proline-rich peptides against S. aureus, showing a synergistic effect (Chernysh et al., 2018). To our knowledge, for the G– bacteria, only one literature reported that clindamycin combined with the peptidomimetic 26 against K. pneumoniae ST258 showed a synergistic effect (Baker et al., 2019); however, no any related studies on the combined use of clindamycin and AMPs against P. aeruginosa have been reported. There are three main types of bacterial resistance mechanisms to clindamycin as reported, including MLSb resistance, mutations in ribosome binding sites, and active efflux of antibiotics from the periplasmic space (Spizek et al., 2004), which mainly occurs in Gram-negative bacteria (Leclercq and Courvalin, 1991). As mentioned in the Introduction, the transmembrane efflux pumps are also considered be the cause of the intrinsic resistance of P. aeruginosa, through which the antibiotics can be effectively taken out of the bacteria (Li et al., 1994). The concentration of clindamycin used in the study was 25 μg·ml−1, which is much lower than the MIC of clindamycin against P. aeruginosa (1,000 μg·ml−1). At the same time, in the synergistic system, only 1/2 MIC of the AMPs were used. At this concentration, the membrane of P. aeruginosa might not be completely disrupted, so the transmembrane efflux pumps of P. aeruginosa might still have a certain effect, and the intracellular concentration of the antibiotics might be reduced by the transmembrane efflux pumps of P. aeruginosa. As a result, the intracellular clindamycin cannot effectively inhibit the synthesis of bacterial protein. Therefore, the combination of clindamycin and the AMPs had no obvious synergistic effect on P. aeruginosa.
In the clinic, penicillin, ceftizoxime, cefotiam, and vancomycin are usually used to treat Gram-positive bacterial infections. Penicillin, ceftizoxime, and cefotiam are all β-lactam antibiotics. They can inhibit peptide bond formation by competitively binding penicillin binding proteins (PBP), prevent the cross-linking of peptidoglycan units, and inhibit the synthesis of bacteria cell walls (Wise and Park, 1965; Waxman et al., 1980). In addition, vancomycin is a glycopeptide antibiotic that can specifically bind to D-Ala-D-Ala dipeptide of the peptidoglycan intermediates, inhibit transglycosylation and/or transpeptidation, overall weaken the peptidoglycan layers, and make the bacterial cells susceptible to changes in osmotic pressure, sequentially inducing cell lysis (Kahne et al., 2005). For the bacterial cell walls, there are three main layers on the cell walls of Gram-negative bacteria, including the outer membrane (OM), the peptidoglycan cell wall, and the cytoplasmic or inner membrane (IM) (Glauert and Thornley, 1969). The outer membrane is mainly composed of LPS (Kamio and Nikaido, 1976), which can protect Gram-negative bacteria from environmental influences by excluding toxic molecules and providing an extra stable layer around the cell. Compared with Gram-positive bacteria, the peptidoglycan layer of Gram-negative bacterial cells is relatively thin. The peptidoglycan layer in the cell walls of P. aeruginosa is only 2.41 ± 0.54 nm thick (Matias et al., 2003), while the Gram-positive bacteria lack the outer membrane and are surrounded by the peptidoglycan layers that are several times thicker than Gram-negative bacteria. The thickness of those peptidoglycan layers ranges from 30 to 100 nm (Silhavy et al., 2010).
For the Gram-negative bacteria P. aeruginosa, the outer membrane would act as a potential barrier to the entrance of antibiotics. In the study, when the antibiotics were used in combination with AMPs Sphistin or Sph12−38, the membrane perturbations caused by the AMPs might allow more antibiotics to enter the bacterial cells. Nevertheless, since the main target for the β-lactam antibiotics (like three antibiotics in the study) and vancomycin is peptidoglycan synthesis, even if the membrane perturbations accelerated the entry of these four antibiotics into bacteria and possibly affect the peptidoglycan synthesis of P. aeruginosa, their action could not affect the integrity of the outer membrane. In addition, a low concentration of the AMPs (<1/2 MIC) could not completely destroy the structure of the outer membrane, and the cell morphology of P. aeruginosa can maintain relative integrity, indicating that the bacteria remained alive. Therefore, when these two antibiotics were used in combination with the AMPs against P. aeruginosa, no significant synergistic effect was observed.
The in vitro antibacterial tests indicated that the combination of Sphistin with rifampicin and azithromycin killed the pathogens efficiently. To demonstrate the synergy effects further, we tested the antibacterial efficiency in vivo. Similar to the experimental results in vitro, the remarkable effect appeared using Sph12−38 in combination with rifampicin that promoted the wound healing significantly (Figures 6A,B), whereas no significant effect was found using Sph12−38 or rifampicin alone. The underlying mechanism was presumed as follows. The AMPs could induce the permeabilization of bacterial cells, facilitating rifampicin to access the cells and bind their binding sites; alternatively, the peptides stimulate the immune systems of the host and then rifampicin could play an antibacterial role independently of the AMPs (Vaara and Porro, 1996; Yeaman and Yount, 2003; Balakrishna et al., 2006). Nevertheless, unlike rifampicin, azithromycin alone or in combination with Sph12−38 significantly facilitated the wound healing. Otherwise, Sphistin/Sph12−38 could bind to LPS and permeabilize the bacterial membrane; when combined with rifampicin and azithromycin, Sphistin/Sph12−38 promoted the intracellular uptake of the antibiotics and subsequently enhanced the bactericidal activity of both agents against P. aeruginosa. Although P. aeruginosa was non-susceptible to rifampicin or azithromycin, when in combination with Sphistin/Sph12−38, they all showed higher antibacterial efficiency; the combination of Sphistin/Sph12−38 with rifampicin and azithromycin might be potentially used for the prevention and treatment of infections caused by P. aeruginosa; however, more work needs to be done in the future.
Data Availability Statement
The raw data supporting the conclusions of this article will be made available by the authors, without undue reservation.
Ethics Statement
The animal study was reviewed and approved by Xiamen University Laboratory Animal Management and Ethics Committee.
Author Contributions
K-JW design the research work, supervised, and revised the manuscript. JL performed the experiment, co-designed, and wrote the paper. XW and HZ participated the antibacterial experiment and the transmission electron microscopy (TEM) experiment, and verified the overall replication/reproducibility of results/experiments and other research outputs. HP provided the antimicrobial peptides and co-designed the in vivo experiment. K-JW and FC supervised and revised the research work. All authors contributed to the article and approved the submitted version.
Funding
This work was sponsored by the National Natural Science Foundation of China (grant # 41676158 and 41806162); the Fundamental Research Funds for the Central Universities (grant # 20720190109 and 20720180100); and the Fujian Marine Economic Development Subsidy Fund Project from the Fujian Ocean and Fisheries Department (grant # FJHJF-L-2019-1). The National Natural Science Foundation of China (NSFC) is in charge of the administration of NSFC in accordance with the law and operates relatively independently. It is responsible for the organization and implementation of subsidy plans, project setup and review, project approval and supervision. The Fundamental Research Funds for the Central Universities funded by Ministry of Education of the People's Republic of China. Fujian Marine Economic Development Subsidy Fund Project from the Fujian Ocean and Fisheries Department. The funds received for open access publication fees from Xiamen University. This study also received funding from Xiamen Science and Technology Planning Project (grant # 3502Z20203012).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
References
Alvarez-Elcoro, S., and Enzler, M. J. (1999). The macrolides: erythromycin, clarithromycin, and azithromycin. Mayo Clin. Proc. 74, 613–34. doi: 10.4065/74.6.613
Andersson, D. I., and Levin, B. R. (1999). The biological cost of antibiotic resistance, Curr. Opin. Microbiol. 2, 489–493. doi: 10.1016/S1369-5274(99)00005-3
Arii, K., Kawada-Matsuo, M., Oogai, Y., Noguchi, K., and Komatsuzawa, H. (2019). Single mutations in BraRS confer high resistance against nisin A in Staphylococcus aureus. Microbiologyopen 8:e791. doi: 10.1002/mbo3.791
Baker, K. R., Jana, B., Hansen, A. M., Nielsen, H. M., Franzyk, H., and Guardabassi, L. (2019). Repurposing azithromycin and rifampicin against gram-negative pathogens by combination with peptidomimetics. Front. Cell. Infect. Microbiol. 9:236. doi: 10.3389/fcimb.2019.00236
Balakrishna, R., Wood, S. J., Nguyen, T. B., Miller, K. A., Kumar, E. S., Datta, A., et al. (2006). Structural correlates of antibacterial and membrane-permeabilizing activities in acylpolyamines. Antimicrob. Agents Chemother. 50, 852–861. doi: 10.1128/AAC.50.3.852-861.2006
Band, V. I., Hufnagel, D. A., Jaggavarapu, S., Sherman, E. X., Wozniak, J. E., Satola, S. W., et al. (2019). Antibiotic combinations that exploit heteroresistance to multiple drugs effectively control infection. Nat. Microbiol. 4, 1627–1635. doi: 10.1038/s41564-019-0480-z
Bliziotis, I., Ntziora, F., Lawrence, K., and Falagas, M. (2007). Rifampin as adjuvant treatment of Gram-positive bacterial infections: a systematic review of comparative clinical trials. Eur. J. Clin. Microbiol. Infect. Dis. 26:849. doi: 10.1007/s10096-007-0378-1
Blonder, J., Ghose, M. B., Xiao, W., Camp, D. G., Wingred, M., Davis, R. W., et al. (2004). Global analysis of the membrane subproteome of pseudomonas aeruginosa using liquid chromatography-tandem mass spectrometry. J. Proteome Res. 3, 434–444. doi: 10.1021/pr034074w
Brogden, K. A. (2005). Antimicrobial peptides: pore formers or metabolic inhibitors in bacteria? Nat. Rev. Microbiol. 3, 238–50. doi: 10.1038/nrmicro1098
Brogden, K. A., Ackermann, M., McCray, P. B., and Tack, B. F. (2003). Antimicrobial peptides in animals and their role in host defences. Int. J. Antimicrob. Agents 22, 465–478. doi: 10.1016/S0924-8579(03)00180-8
Cai, Y., Yang, D., Wang, J., and Wang, R. (2018). Activity of colistin alone or in combination with rifampicin or meropenem in a carbapenem-resistant bioluminescent Pseudomonas aeruginosa intraperitoneal murine infection model. J. Antimicrob. Chemother. 73, 456–461. doi: 10.1093/jac/dkx399
Campbell, E. A., Korzheva, N., Mustaev, A., Murakami, K., Nair, S., Goldfarb, A., et al. (2001). Structural mechanism for rifampicin inhibition of bacterial rna polymerase. Cell 104, 901–912. doi: 10.1016/S0092-8674(01)00286-0
Cassone, M., and Otvos, L. (2010). Synergy among antibacterial peptides and between peptides and small-molecule antibiotics. Expert. Rev. Anti Infect. Ther. 8, 703–716. doi: 10.1586/eri.10.38
C. D. C. P. (US) Centres for Disease Control and Prevention (US). (2013). Antibiotic resistance threats in the United States, 2013. Centres for Disease Control and Prevention, US Department of Health and Human Services.
Chen, B., Fan, D. Q., Zhu, K. X., Shan, Z. G., Chen, F. Y., Hou, L., et al. (2015). Mechanism study on a new antimicrobial peptide Sphistin derived from the N-terminus of crab histone H2A identified in haemolymphs of Scylla paramamosain. Fish. Shellfish. Immunol. 47, 833–46. doi: 10.1016/j.fsi.2015.10.010
Chernysh, S. I., Gordya, N., Tulin, D., and Yakovlev, A. B. (2018). Biofilm infections between scylla and charybdis: interplay of host antimicrobial peptides and antibiotics. Infect. Drug Resist. 11, 501–514. doi: 10.2147/IDR.S157847
Chongsiriwatana, N. P., Patch, J. A., Czyzewski, A. M., Dohm, M. T., Ivankin, A., Gidalevitz, D., et al. (2008). Peptoids that mimic the structure, function, and mechanism of helical antimicrobial peptides. Proc. Natl. Acad. Sci. U.S.A. 105, 2794–2799. doi: 10.1073/pnas.0708254105
Cirioni, O., Ghiselli, R., Orlando, F., Silvestri, C., Mocchegiani, F., Rocchi, M., et al. (2007). Efficacy of colistin/rifampin combination in experimental rat models of sepsis due to a multiresistant Pseudomonas aeruginosa strain. Crit. Care Med. 35, 1717–1723. doi: 10.1097/01.CCM.0000266685.25436.03
Cirioni, O., Silvestri, C., Ghiselli, R., Orlando, F., Riva, A., Mocchegiani, F., et al. (2008). Protective effects of the combination of α-helical antimicrobial peptides and rifampicin in three rat models of Pseudomonas aeruginosa infection. J. Antimicrob. Chemother. 62, 1332–1338. doi: 10.1093/jac/dkn393
C. L. S. Institute. (2012). Methods for Dilution Antimicrobial Susceptibility Tests for Bacteria That Grow Aerobically; Approved Standards—Ninth Edition (Wayne: CLSI document M07-A9).
Dobson, A. J., Purves, J., Kamysz, W., and Rolff, J. (2013). Comparing selection on s. aureus between antimicrobial peptides and common antibiotics. PLOS ONE, 8:e0076521. doi: 10.1371/journal.pone.0076521
El Shazely, B., Yu, G. Z., Johnston, P. R., and Rolff, J. (2020). Resistance evolution against antimicrobial peptides in staphylococcus aureus alters pharmacodynamics beyond the MIC. Front. Microbiol. 11:103. doi: 10.3389/fmicb.2020.00103
Evans, M. E., Feola, D. J., and Rapp, R. P. (1999). Polymyxin B sulfate and colistin: old antibiotics for emerging multiresistant gram-negative bacteria. Ann. Pharmacother. 33, 960–967. doi: 10.1345/aph.18426
French, G. L. (2010). The continuing crisis in antibiotic resistance. Int. J. Antimicrob. Agents 36, S3–S7. doi: 10.1016/S0924-8579(10)70003-0
Frieden, T. (2013). Antibiotic resistance Threats in the United States, 2013, Vol. 23. Centers for Disease Control and Prevention, US Department of Health and Human Services, 11–28.
Gardner, T. B., and Hill, D. R. (2001). Treatment of giardiasis. Clin. Microbiol. Rev. 14, 114–128. doi: 10.1128/CMR.14.1.114-128.2001
Giamarellos-Bourboulis, E., Sambatakou, H., Galani, I., and Giamarellou, H. (2003). In vitro interaction of colistin and rifampin on multidrug-resistant Pseudomonas aeruginosa. J. Chemother. 15, 235–238. doi: 10.1179/joc.2003.15.3.235
Glauert, A. M., and Thornley, M. J. (1969). The topography of the bacterial cell wall. Ann. Rev. Microbiol. 23, 159–198. doi: 10.1146/annurev.mi.23.100169.001111
Guay, D. R. J. D. (2008). Contemporary management of uncomplicated urinary tract infections. Drugs. 68, 1169–1205. doi: 10.2165/00003495-200868090-00002
Habets, M. G. J. L., Rozen, D. E., and Brockhurst, M. A. (2012). Variation in Streptococcus pneumoniae susceptibility to human antimicrobial peptides may mediate intraspecific competition. Proc. Biol. Sci. 279, 3803–3811. doi: 10.1098/rspb.2012.1118
Hancock, R. E. (1998). Resistance mechanisms in Pseudomonas aeruginosa and other nonfermentative gram-negative bacteria. Clin. Infect. Dis. 27, S93–S99 doi: 10.1086/514909
Hancock, R. E., and Speert, D. P. (2000). Antibiotic resistance in Pseudomonas aeruginosa: mechanisms and impact on treatment. Drug Resist. Updat. 3, 247–255. doi: 10.1054/drup.2000.0152
Haney, E. F., Mansour, S. C., and Hancock, R. E. (eds.). (2017). “Antimicrobial peptides: an introduction,” in Antimicrobial Peptides (New York, NY: Springer) doi: 10.1007/978-1-4939-6737-7_1
Heinz Floss, G., and Yu, W.-T. (2005). Rifamycin-mode of action, resistance, and biosynthesis. Chem. Rev. 105, 621–32. doi: 10.1021/cr030112j
Iwagaki, A., Porro, M., and Pollack, M. (2000). Influence of synthetic antiendotoxin peptides on lipopolysaccharide (LPS) recognition and LPS-induced proinflammatory cytokine responses by cells expressing membrane-bound CD14. Infect. Immun. 68, 1655–1663. doi: 10.1128/IAI.68.3.1655-1663.2000
Kahne, D., Leimkuhler, C., Lu, W., and Walsh, C. T. (2005). Glycopeptide and lipoglycopeptide antibiotics. Chem. Rev. 105, 425–448. doi: 10.1021/cr030103a
Kamio, Y., and Nikaido, H. (1976). Outer membrane of salmonella typhimurium: accessibility of phospholipid head groups to phospholipase c and cyanogen bromide activated dextran in the external medium. Biochemistry 15, 2561–2570. doi: 10.1021/bi00657a012
Khara, J. S., Lim, F. K., Wang, Y., Ke, X.-Y., Voo, Z. X., Yang, Y. Y., et al. (2015). Designing α-helical peptides with enhanced synergism and selectivity against mycobacterium smegmatis: discerning the role of hydrophobicity and helicity. Acta Biomater. 28, 99–108 doi: 10.1016/j.actbio.2015.09.015
Khara, J. S., Wang, Y., Ke, X. Y., Liu, S. Q., Newton, S. M., Langford, P. R., et al. (2014). Anti-mycobacterial activities of synthetic cationic alpha-helical peptides and their synergism with rifampicin. Biomaterials, 35, 2032–2038. doi: 10.1016/j.biomaterials.2013.11.035
Koppen, B. C., Mulder, P. P., G., de Boer, L., Riool, M., Drijfhout, J. W., et al. (2019). Synergistic microbicidal effect of cationic antimicrobial peptides and teicoplanin against planktonic and biofilm-encased Staphylococcus aureus. Int. J. Antimicrob. Agents 53, 143–151. doi: 10.1016/j.ijantimicag.2018.10.002
Koshlukova, S. E., Lloyd, T. L., Araujo, M. W., B., and Edgerton, M. (1999). Salivary histatin 5 induces non-lytic release of ATP from Candida albicans leading to cell death. J. Biol. Chem. 274, 18872–18879. doi: 10.1074/jbc.274.27.18872
Leclercq, R., and Courvalin, P. (1991). Bacterial resistance to macrolide, lincosamide, and streptogramin antibiotics by target modification. Antimicrob. Agents Chemother. 35, 1267–1272 doi: 10.1128/AAC.35.7.1267
Levin, B., Lipsitch, M., Perrot, V., Schrag, S., Antia, R., Simonsen, L., et al. (1997). The population genetics of antibiotic resistance. Clin. Infect. Dis. 24, S9-S16. doi: 10.1093/clinids/24.Supplement_1.S9
Lew, W., Pai, M., Oxlade, O., Martin, D., and Menzies, D. J. A. (2008). Initial drug resistance and tuberculosis treatment outcomes: systematic review and meta-analysis, Ann. Intern. Med. 149, 123–134. doi: 10.7326/0003-4819-149-2-200807150-00008
Li, D., Yang, Y., Tian, Z., Lv, J., Sun, F., Wang, Q., et al. (2017). Synergistic antibiotic effect of looped antimicrobial peptide CLP-19 with bactericidal and bacteriostatic agents. Oncotarget 8, 55958–55966. doi: 10.18632/oncotarget.18124
Li, X. Z., Livermore, D. M., and Nikaido, H. (1994). Role of efflux pump(s) in intrinsic resistance of Pseudomonas aeruginosa: resistance to tetracycline, chloramphenicol, and norfloxacin. Antimicrob Agents Chemother, 38, 1732–1741 doi: 10.1128/AAC.38.8.1732
Livermore, D. M. (2002). Multiple mechanisms of antimicrobial resistance in Pseudomonas aeruginosa: our worst nightmare? Clin. Infect. Dis. 34, 634–640. doi: 10.1086/338782
Lyczak, J. B., Cannon, C. L., and Pier, G. B. (2000). Establishment of Pseudomonas aeruginosa infection: lessons from a versatile opportunist. Microbes Infect 2, 1051–60. doi: 10.1016/S1286-4579(00)01259-4
Ma, X. W., Hou, L., Chen, B., Fan, D. Q., Chen, Y. C., Yang, Y., et al. (2017). A truncated Sph12-38 with potent antimicrobial activity showing resistance against bacterial challenge in Oryzias melastigma. Fish. Shellfish Immunol. 67, 561–570. doi: 10.1016/j.fsi.2017.06.013
Magiorakos, A. P., Srinivasan, A., Carey, R. B., Carmeli, Y., Falagas, M. E., Giske, C. G., et al. (2012). Multidrug-resistant, extensively drug-resistant and pandrug-resistant bacteria: an international expert proposal for interim standard definitions for acquired resistance. Clinical Microbiol. Infect. 18, 268–281. doi: 10.1111/j.1469-0691.2011.03570.x
Makarova, O., Johnston, P. R., Rodriguezrojas, A., Shazely, B. E., Morales, J. M., and Rolff, J. (2018). Genomics of experimental adaptation of Staphylococcus aureus to a natural combination of insect antimicrobial peptides. Sci. Rep. 8:15359 (2018) doi: 10.1038/s41598-018-33593-7
Matias, V. R., F., Alamoudi, A., Dubochet, J., and Beveridge, T. J. (2003). Cryo-Transmission Electron Microscopy of Frozen-Hydrated Sections of Escherichia coli and Pseudomonas aeruginosa. J. Bacteriol. 185, 6112–6118. doi: 10.1128/JB.185.20.6112-6118.2003
Mazzei, T., Mini, E., Novelli, A., and Periti, P. (1993). Chemistry and mode of action of macrolides. J. Antimicrob. Chemother. 31(Suppl C), 1–9 doi: 10.1093/jac/31.suppl_C.1
Menninger, J. R., and Otto, D. P. (1982). Erythromycin, carbomycin, and spiramycin inhibit protein synthesis by stimulating the dissociation of peptidyl-tRNA from ribosomes. Antimicrob. Agents Chemother. 21, 811–818. doi: 10.1128/AAC.21.5.811
Nguschwemlein, M., Dumond, J., Rudd, L., and Rigaud, J. (2014). In vitro synergy between some cationic amphipathic cyclooctapeptides and antibiotics. Aust. J. Chem. 68, 218–223. doi: 10.1071/CH14427
Nord, C. E., and Kager, L. (1983). Tinidazole - microbiology, pharmacology and efficacy in anaerobic infections. Infection 11, 54–60. doi: 10.1007/BF01651361
Odds, F. C. (2003). Synergy, antagonism, and what the chequerboard puts between them. J. Antimicrob. Chemother. 52, 1–1. doi: 10.1093/jac/dkg301
Pacios, O., Blasco, L., Bleriot, I., Fernandezgarcia, L., Bardanca, M. G., Ambroa, A., et al. (2020). Strategies to combat multidrug-resistant and persistent infectious diseases. J. Antib. 9:65 doi: 10.3390/antibiotics9020065
Pal, C., Papp, B., and Lazar, V. (2015). Collateral sensitivity of antibiotic-resistant microbes. Trends Microbiol. 23, 401–407. doi: 10.1016/j.tim.2015.02.009
Pankey, G., Ashcraft, D., and Patel, N. (2005). In vitro synergy of daptomycin plus rifampin against Enterococcus faecium resistant to both linezolid and vancomycin. Antimicrob. Agents Chemother. 49, 5166–5168 doi: 10.1128/AAC.49.12.5166-5168.2005
Pankey, G. A., and Ashcraft, D. S. (2005). In vitro synergy of ciprofloxacin and gatifloxacin against ciprofloxacin-resistant Pseudomonas aeruginosa. Antimicrob. Agents Chemother. 49, 2959–2964. doi: 10.1128/AAC.49.7.2959-2964.2005
Peters, D. H., Friedel, H. A., and McTavish, D. (1992). azithromycin - a review of its antimicrobial activity, pharmacokinetic properties and clinical efficacy Drugs 44, 750–799. doi: 10.2165/00003495-199244050-00007
Petersen, P. J., Labthavikul, P., Jones, C. H., and Bradford, P. A. (2006). In vitro antibacterial activities of tigecycline in combination with other antimicrobial agents determined by chequerboard and time-kill kinetic analysis. J. Antimicrob. Chemother. 57, 573–576. doi: 10.1093/jac/dki477
Raether, W., and Hanel, H. (2003). Nitroheterocyclic drugs with broad spectrum activity. Parasitol. Res. 90(Supp 1), S19–39. doi: 10.1007/s00436-002-0754-9
Rand, K. H., Houck, H. J., Brown, P., and Bennett, D. (1993). Reproducibility of the microdilution checkerboard method for antibiotic synergy. Antimicrob. Agents Chemother. 37, 613–615. doi: 10.1128/AAC.37.3.613
Retsema, J., Girard, A., Schelkly, W., Manousos, M., Anderson, M., Bright, G., et al. (1987). Spectrum and mode of action of azithromycin (CP-62,993), a new 15-membered-ring macrolide with improved potency against gram-negative organisms. Antimicrob. Agents Chemother. 31, 1939–47. doi: 10.1128/AAC.31.12.1939
Roemhild, R., and Schulenburg, H. (2019). Evolutionary ecology meets the antibiotic crisis: can we control pathogen adaptation through sequential therapy? Evol. Med. Public Health 2019, 37–45. doi: 10.1093/emph/eoz008
Scott, M. G., Yan, H., and Hancock, R. E. (1999). Biological properties of structurally related α-helical cationic antimicrobial peptides. Infect. Immun. 67, 2005–2009. doi: 10.1128/IAI.67.4.2005-2009.1999
Shai, Y. (2002). Mode of action of membrane active antimicrobial peptides. Biopolymers 66, 236–48. doi: 10.1002/bip.10260
Silhavy, T. J., Kahne, D., and Walker, S. (2010). The Bacterial Cell Envelope. Cold Spring Harbor Perspec. Biol. 2:414. doi: 10.1101/cshperspect.a000414
Spizek, J., Novotna, J., and Rezanka, T. (2004). Lincosamides: chemical structure, biosynthesis, mechanism of action, resistance, and applications. Adv. Appl. Microbiol. 56, 121–154 doi: 10.1016/S0065-2164(04)56004-5
Tasca, T., Borges, F. P., Bonan, C. D., De Carli, G. A., Battastini, A. M. O., et al. (2003). Effects of metronidazole and tinidazole on NTPDase1 and ecto-5'-nucleotidase from intact cells of Trichomonas vaginalis. Fems Microbiol. Lett. 226, 379–384. doi: 10.1016/S0378-1097(03)00637-2
Tascini, C., Gemignani, G., Ferranti, S., Tagliaferri, E., Leonildi, A., Lucarini, A., et al. (2004). Microbiological activity and clinical efficacy of a colistin and rifampin combination in multidrug-resistant Pseudomonas aeruginosa infections. J. Chemother. 16, 282–287. doi: 10.1179/joc.2004.16.3.282
Tenover, F. C. (2006). Mechanisms of antimicrobial resistance in bacteria. Am. J. Med. 119, S3-S10 doi: 10.1016/j.amjmed.2006.03.011
Timurkaynak, F., Can, F., Azap, Ö. K., Demirbilek, M., Arslan, H., and Karaman, S. Ö. (2006). In vitro activities of non-traditional antimicrobials alone or in combination against multidrug-resistant strains of Pseudomonas aeruginosa and Acinetobacter baumannii isolated from intensive care units. Int. J. Antimicrob. Agents 27, 224–228. doi: 10.1016/j.ijantimicag.2005.10.012
Vaara, M. (1993). Outer membrane permeability barrier to azithromycin, clarithromycin, and roxithromycin in gram-negative enteric bacteria. Antimicrob. Agents Chemother. 37, 354–356. doi: 10.1128/AAC.37.2.354
Vaara, M., and Porro, M. (1996). Group of peptides that act synergistically with hydrophobic antibiotics against gram-negative enteric bacteria. Antimicrob. Agents Chemother. 40, 1801–1805. doi: 10.1128/AAC.40.8.1801
Ventola, C. L. (2015). The antibiotic resistance crisis: part 1: causes and threats. Pharm. Therap. 40, 277−83.
Vizioli, J., and Salzet, M. (2002). Antimicrobial peptides from animals: focus on invertebrates. Trends Pharmacol. Sci. 23, 494–6. doi: 10.1016/S0165-6147(02)02105-3
Wade, D., Boman, A., Wåhlin, B., Drain, C., Andreu, D., Boman, H. G., et al. (1990). All-D amino acid-containing channel-forming antibiotic peptides. Proc. Natl. Acad. Sci.U.S.A. 87, 4761–4765. doi: 10.1073/pnas.87.12.4761
Walter, W., and Staehelin, M. (1971). Actions of the Rifamycins. Bacteriol. Rev. 35:290. doi: 10.1128/MMBR.35.3.290-309.1971
Waxman, D. J., Yocum, R. R., and Strominger, J. L. (1980). Penicillins and cephalosporins are active site-directed acylating agents - evidence in support of the substrate-analog hypothesis. Philos. Trans. Roy. Soc. B Biol. Sci. 289, 257–271. doi: 10.1098/rstb.1980.0044
Wise, E. M., and Park, J. T. (1965). Penicillin: its basic site of action as an inhibitor of a peptide cross-linking reaction in cell wall mucopeptide synthesis. Proc. Natl. Acad. Sci.U.S.A. 54, 75–81. doi: 10.1073/pnas.54.1.75
Woodford, N., and Livermore, D. M. J. J. o. I. (2009). Infections caused by Gram-positive bacteria: a review of the global challenge. J. Infect. 59, S4-S16. doi: 10.1016/S0163-4453(09)60003-7
Xu, J. C., Duan, X. M., Wu, H., and Zhou, Q. (2013). Surveillance and Correlation of Antimicrobial Usage and Resistance of Pseudomonas aeruginosa: A Hospital Population-Based Study. PLoS ONE 8:7 doi: 10.1371/journal.pone.0078604
Yamamoto, N., and Tamura, A. (2014). Designing cell-aggregating peptides without cytotoxicity. Biomacromolecules 15, 512–523 (2014) doi: 10.1021/bm4014414
Yang, D., Ni, W., Wang, R., and Wang, J. (2016). In vitro activity of antibiotic combination against carbapenems resistant Pseudomonas aeruginosa. Chin. J. Clin. Pharmacol. 32, 2269–72. doi: 10.1590/0037-8682-0012-2013
Yeaman, M. R., and Yount, N. Y. (2003). Mechanisms of antimicrobial peptide action and resistance. Pharmacol. Rev. 55, 27–55. doi: 10.1124/pr.55.1.2
Yethon, J., and Whitfield, C. (2001). Lipopolysaccharide as a target for the development of novel therapeutics in gram-negative bacteria. Curr. Drug Targets Infect. Disord. 1, 91–106. doi: 10.2174/1568005014606143
Yu, G., Baeder, D. Y., Regoes, R. R., and Rolff, J. (2018). Predicting drug resistance evolution: insights from antimicrobial peptides and antibiotics. Proc. Roy. Soc. B Biol. Sci. 285:20172687 doi: 10.1098/rspb.2017.2687
Zasloff, M. (2002). Antimicrobial peptides of multicellular organisms. Nature 415, 389–95. doi: 10.1038/415389a
Zhang, X. L., Gu, B. N., Mei, Y. N., Wen, Y., and Xia, W. Y. (2015). Increasing resistance rate to carbapenem among blood culture isolates of Klebsiella pneumoniae, Acinetobacter baumannii and Pseudomonas aeruginosa in a university-affiliated hospital in China, 2004–2011. J. Antib. 68, 115–120. doi: 10.1038/ja.2014.119
Keywords: antimicrobial peptides, Sphistin, Sph12−38, Pseudomonas aeruginosa, rifampicin, azithromycin, synergistic efficacy
Citation: Liu J, Chen F, Wang X, Peng H, Zhang H and Wang K-J (2020) The Synergistic Effect of Mud Crab Antimicrobial Peptides Sphistin and Sph12−38 With Antibiotics Azithromycin and Rifampicin Enhances Bactericidal Activity Against Pseudomonas Aeruginosa. Front. Cell. Infect. Microbiol. 10:572849. doi: 10.3389/fcimb.2020.572849
Received: 15 June 2020; Accepted: 02 September 2020;
Published: 23 October 2020.
Edited by:
Mathew Upton, University of Plymouth, United KingdomReviewed by:
Keith Miller, Sheffield Hallam University, United KingdomRoderich Roemhild, Uppsala University, Sweden
Copyright © 2020 Liu, Chen, Wang, Peng, Zhang and Wang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Ke-Jian Wang, wkjian@xmu.edu.cn
 Jie Liu
Jie Liu Fangyi Chen
Fangyi Chen Xiaofei Wang1
Xiaofei Wang1  Hui Peng
Hui Peng Hua Zhang
Hua Zhang Ke-Jian Wang
Ke-Jian Wang