Abstract
Standardized approaches to measuring clinical utility will enable more robust evaluations of genetic tests. To characterize how clinical utility has been measured, this scoping review examined outcomes used to operationalize this concept in the context of genetic testing, spanning relevant literature (2015–2017). The search strategy and analysis were guided by the Fryback and Thornbury hierarchical model of efficacy (FT Model). Through searches in Ovid MEDLINE, EMBASE and Web of Science, 194 publications were identified for inclusion. Two coders reviewed titles, abstracts, and full texts to determine eligibility. Results were analyzed using thematic and frequency analyses. This review generated a catalog of outcomes mapped to the efficacy domains of the FT Model. The degree of representation observed in each domain varied by the clinical purpose and clinical indication of genetic testing. Diagnostic accuracy (68%), technical (28.4%), and patient outcome (28.4%) efficacy studies were represented at the highest rate. Findings suggest that the FT Model is suitable for the genetics context however domain refinements may be warranted. More diverse clinical settings, robust study designs, and novel strategies for measuring clinical utility are needed.
Similar content being viewed by others
Introduction
Advances in genomic medicine have generated much enthusiasm for early disease detection, individualized treatment, and optimized health outcomes [1, 2]. While evidence is compelling related to the analytic performance of emerging genomic technologies, pressures from evidentiary review bodies, policymakers, and payors to define and measure the clinical and economic value of genetic testing are increasing [3, 4]. Frameworks for evaluating genetic tests such as those used by the Evaluation of Genomic Applications in Practice and Prevention (EGAPP™) Working Group and the United States Preventive Services Task Force have been in use for many years and have guided test implementation decisions and clinical practice [5]. Within these evaluative frameworks, the value or ‘clinical utility’ parameter has been defined in various ways. For example, the much-endorsed ACCE framework for evaluating genetic tests (i.e., Analytic validity, Clinical validity, Clinical utility, Ethical, legal, and social implications), defines clinical utility with respect to whether genetic testing improves patient outcomes and/or adds value to the clinical decision-making process [6,7,8]. Recently, the American College of Medical Genetics and Genomics (ACMG) and other organizations have called for an even more expansive definition of this concept [9], arguing that assessments of clinical utility should attend to the effect of genetic testing on diagnosis, prognosis, therapeutic management, the health and psychological well-being of patients and their relatives, and health care system costs [6, 8,9,10,11].
Lack of consensus on how to operationalize the notion of clinical utility can result in inadequate or inconsistent determinations of value across laboratories, hospitals, professional societies, and payors, in turn leading to variations in test quality, coverage, and access [12]. In part, this lack of consensus relates to the challenge of capturing the indirect relationship between a test result and a specific clinical outcome [13]. In contrast with therapeutics that are hypothesized to act directly on health outcomes, a diagnostic test result may inform clinical decision making which may in turn, impact outcomes related to disease burden and patient experience over time [14, 15]. As such, it has been argued that a “chain of evidence” must be applied to measuring the value of genetic testing; intermediate links in a chain of evidence, consisting of surrogate outcomes (i.e., measures that impact or correlate with clinical outcomes) are essential to characterizing a more expansive notion of clinical utility [7, 13,14,15,16].
Given the current emphasis on defining and measuring the notion of value in the context of genomic medicine and the rate at which new test applications are entering clinical practice, a comprehensive understanding of the body of empiric work that reflects on clinical utility as a core component of genetic service delivery can serve as an important step towards more consistently defining and measuring this concept [8, 12]. The FT Model is widely used for the evaluation of medical tests and the suitability of the FT Model for use in the genetic testing context has been explored previously [5, 8]. The hierarchical nature of the framework is well-suited to the context of genetics because the components of efficacy are specific, well defined, and linked as a chain of evidence [8, 16, 17]. Given these characteristics, the FT Model provides a useful structure for organizing literature related to the clinical utility of genetic testing [8, 16]. This was illustrated in the modified FT Model developed by the National Academies of Sciences, Engineering, and Medicine (NAS) in which genetic test assessment methods were integrated with relevant health outcomes previously identified by the EGAPP working group [8]. Using the Fryback and Thornbury hierarchical model of efficacy (FT Model) as a conceptual framework, the objective of this scoping review was to explore the range of outcomes used, in peer-reviewed literature, to measure the clinical utility of genetic testing in a range of clinical settings.
Materials and methods
Search Strategy Aligned with Arksey and O’Malley’s methodology [17] and the Preferred Reporting Items for Systematic Reviews and Meta-Analyses extension for Scoping Reviews (PRISMA-ScR), a review of empiric literature that assesses the clinical utility of genetic testing was conducted. The FT Model was used to guide the search strategy. It consists of six domains of efficacy: technical efficacy (i.e., laboratory performance), diagnostic accuracy efficacy (i.e., clinical sensitivity and specificity), diagnostic thinking efficacy (i.e., impact on clinician’s diagnostic process), therapeutic efficacy (i.e., impact on clinical management), patient outcome efficacy (i.e., patient benefit) and societal outcome efficacy (i.e., cost-benefit, cost-effectiveness, societal acceptability) [8, 16]. For the purpose of this review, clinical utility was defined to include diagnostic thinking efficacy, therapeutic efficacy and patient outcome efficacy. MeSH and search terms related to these concepts included but were not limited to clinical decision-making, clinical management, diagnostic value, treatment outcome, patient outcome, mortality rate, morbidity, and quality of life. Studies were identified through searches in Ovid MEDLINE, EMBASE, and Web of Science. Given the importance of the broad definition of clinical utility that was articulated by the ACMG in 2015 [9], we decided to focus on studies published from January 2015 to May 2017 to gauge the alignment of ACMG’s conception of clinical utility with empiric work reported around the same timepoint. The Medline search strategy is described in Table S1.
Article selection
Eligibility criteria were established through an iterative process of article review and consultation with co-authors. Included studies were English, peer-reviewed, and experimental or observational in design. Since the health technology assessment process largely relies on the peer-reviewed literature as its evidence base, this data source was deemed the most relevant for the purposes of our review. While the search strategy focused on specific domains of clinical utility (i.e., diagnostic thinking efficacy, therapeutic efficacy, and patient outcome efficacy), articles identified in the search that also reflected on the utility of genetic testing from a laboratory perspective (i.e., technical and diagnostic accuracy efficacy domains) were also included. In addition, articles that reflected on the societal perspective (i.e., societal efficacy domain) in the form of economic evaluations were retained to reflect a full representation of the measures used to assess the value of genetic testing. Conference materials, editorials, consensus statements, literature reviews, practice guidelines, case reports, case series, and descriptions of disease phenotype, genotype or new disease genes were excluded. Exclusions were identified using both EndNote X7-7.1 filters and manual review. Abstracts and full-text articles were reviewed for inclusion by two independent reviewers. To develop consensus and improve consistency, the reviewers met early in the selection phase to compare results and refine selection criteria. To monitor consistency with inclusion decisions, a comparison of the first 1000 abstracts showed that independent reviewers agreed 85.3% of the time. Reviewers agreed 83.6% of the time during the full article review phase. Discrepancies were discussed until consensus was achieved. If consensus proved challenging, a third reviewer was consulted. Results were tracked and managed using EndNote X7-7.1, REDCap, and Microsoft Excel.
Data extraction
The following data elements were extracted from each included article: author(s), publication year, study title, clinical indication for testing (e.g., cancer, cardiovascular disease), genetic test purpose (e.g., diagnosis, screening, clinical management), population type (i.e., adult, pediatric, both), data collection method (i.e., retrospective, prospective, both), study design (i.e., experimental, observational, other), outcome measure(s) used and additional comments. Clinical indication for testing was characterized using the International Statistical Classification of Diseases and Related Health Problems as a guide [18]. Each outcome measure was assigned to an FT Model efficacy domain; this was facilitated by using a modified version of the model [8] that includes analytic questions and suggested measures for each domain. Where an outcome could be assigned to more than one domain, based on the concepts outlined in the FT Model, decision rules were established to navigate domain boundaries. This occurred in three instances: (i) Sensitivity and specificity measures can apply to both the technical efficacy and diagnostic accuracy domains. As such, these measures were assigned to the technical efficacy domain if they were reported in a laboratory setting that focused on the test’s ability to identify a target variant and were assigned to the diagnostic accuracy efficacy domain if they were reported in a clinical context that focused on the test’s ability to identify patients with or without a target disease. (ii) Measures that relate to prognosis can apply to both the diagnostic accuracy and diagnostic thinking efficacy domains. In this review, prognostic outcomes were assigned to the diagnostic thinking efficacy domain because they more closely relate to helping clinicians come to a diagnosis. (iii) Actionable variant detection rate is an outcome measure that can apply to both the diagnostic accuracy and therapeutic efficacy domains. This measure was assigned to the therapeutic efficacy domain because the reviewers determined that it most closely related to medical management. While these decision rules were required for the purpose of data classification, we acknowledge that this classification scheme may not align with how all clinicians would define or use these concepts in clinical practice settings. Outcome measures that did not align with an efficacy domain were categorized as ‘other’. As above, two reviewers extracted data from each included citation and a third reviewer was consulted to resolve discrepancies. Before finalizing the data extraction form, it was pilot tested by the co-authors using a random sample of 50 articles and revised accordingly. The reviewers also met regularly to compare results and ensure a uniform approach to data extraction.
Analysis
Study characteristics were categorized and summarized using frequency tabulations. We tabulated the percentage of studies with outcomes in each efficacy domain for each test purpose and for a subset of the three most common clinical indications for testing. The analysis then focused on characterizing the nature of the outcome measures used in the literature to operationalize the concept of clinical utility. After assigning each measure to an appropriate efficacy domain, thematic categories were established within each domain in order to group related measures together. New thematic categories were added to each efficacy domain until thematic saturation was achieved. For example, within the diagnostic accuracy efficacy domain, the ‘family screening’ category includes cascade testing rate, family member diagnosis rate, and family-member carrier detection rate. Importantly, some of the outcomes listed within each thematic category reflected true validated measures (e.g., State Trait Anxiety Index) and some reflected indicators of specific constructs (e.g., family screening). Finally, we reflected on the suitability of the FT Model as an organizing structure for characterizing this body of evidence.
Results
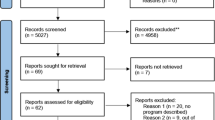
A summary of the scoping review search results is displayed in Fig. 1. A total of 1954 records published between 2015 and 2017 were identified. A total of 1205 abstracts were reviewed following the removal of duplicates and exclusions; 451 studies were selected for full-text review and transferred to a REDCap database. Following full-text review, 194 studies were identified for inclusion.
Study characteristics
Studies related to measuring the clinical utility of genetic testing for cancer were the most common (32.0%), followed by studies related to congenital anomalies (12.4%) and cardiovascular disease (10.8%). The remaining studies were widely distributed across various clinical indications including the nervous system, mental health, and metabolism. Of the studies reviewed, 50.5% involved only adult populations, 21.1% involved adult and pediatric populations, and 17.5% involved only pediatric populations. Most of the included studies used an observational study design (88.7%); randomized (6.7%) and non-randomized (4.6%) experimental designs were used in a minority. Data were collected retrospectively for 50.0%, prospectively for 42.3%, and both prospectively and retrospectively for 8.2%. Diagnosis was the stated clinical purpose for genetic testing in 42.3% followed by clinical management (25.8%), screening (16.5%), and a combination of diagnosis and clinical management (15.5%; Table 1).
Outcomes considered by test purpose and clinical indication
Of the 194 studies reviewed, 28.4% had outcomes that mapped to technical efficacy, 68.0% to diagnostic accuracy efficacy, 1.0% to diagnostic thinking efficacy, 18.6% to therapeutic efficacy, 28.4% to patient outcome efficacy, and 16.5% to societal efficacy (see Supplementary Appendix). Figure 2a presents the proportion of outcome measures assigned to each efficacy domain, by stated test purpose. Outcomes that mapped to the technical and diagnostic accuracy efficacy domains were most common in tests used for diagnosis and/or clinical management. Therapeutic efficacy outcomes were most common in tests with a clinical management purpose. Patient outcome efficacy and societal efficacy outcomes were most common in tests with a clinical management or screening purpose. To explore whether the domains represented in the reviewed literature varied by clinical indication, Fig. 2b presents the proportion of outcome measures assigned to each efficacy domain for the three most common clinical indications for testing: cancer, cardiovascular disease, and congenital anomalies. Across indications, outcomes related to diagnostic accuracy were represented most often while outcomes related to diagnostic thinking were represented least often. In addition, cancer studies used a higher proportion of therapeutic outcomes compared to cardiovascular disease and congenital anomaly studies.
To examine whether the FT model efficacy domains represented in the reviewed literature varied by test purpose or clinical context, study outcomes were organized into thematic groups. a Shows study outcomes organized by efficacy domain and stated test purpose. b Shows study outcomes organized by efficacy domain and the clinical condition under investigation.
Operationalizing the concept of clinical utility
Table 2 presents a sample of the outcome measures identified in each efficacy domain, grouped by thematic category. The percentage of studies within each domain that used measures from the identified thematic categories are reported. For technical efficacy, all of the outcome measures fit within the analytic test performance category, including analytic sensitivity and specificity, sequencing coverage depth, and sequence amplification rate. Within the diagnostic accuracy efficacy domain, we identified four main categories; variant detection rate (e.g., detection rate for variant that affects function, and actionable variant detection rate), clinical diagnosis rate (e.g., diagnosis rate, diagnostic yield/probable-molecular diagnosis rate), clinic test performance (e.g., clinical sensitivity and specificity, positive/negative predictive values) and family screening (e.g., cascade testing rate, family member diagnosis rate). In the diagnostic thinking efficacy domain, impact on diagnostic process was identified as the only thematic category and it included two outcome measures, clinician reported usefulness of testing and the frequency of changed diagnosis/prognosis based on genetic test results. Two categories were identified in the therapeutic efficacy domain. The first category included measures of genetic test impact on clinical recommendations and/or clinical interventions (e.g., percentage of patients that received a revised clinical recommendation following genetic testing) and the second category related to treatment optimization outcomes (e.g., estimated reduction in unnecessary follow-up procedures). Measures in the patient outcome efficacy domain were organized into two categories; health-related indicators (e.g., disease-free interval, disease recurrence rate, quality of life) and behavior indicators (e.g., self-reported changes in health behavior following diabetes genetic testing). Societal outcome efficacy measures were organized into three categories, including cost of testing (e.g., cost per patient, cost per diagnosis, cost per test pathway), cost-effectiveness (e.g., QALYs, ICERs) and cost savings (e.g., mean annualized savings rate associated with test use, incremental cost saving per year). Table S2 presents all measure types identified. Of the 194 studies, 5.2% used outcome measures that could not be mapped to any of the FT Model efficacy domains. These ‘other outcomes’ were organised into four thematic categories; estimated prevalence of disease, parent outcomes, time to diagnosis and genetic test utilization in different clinical settings (see Table S3).
Discussion
The size of the final dataset (n = 194) and the presence of outcome measures in each of the efficacy domains are strong indicators of the breadth of clinical utility-oriented research underway. Capturing this breadth enabled the creation of an empirically grounded and thematically organized catalog of outcome measures that align with the FT Model, a robust framework for evaluating genetic tests. In the studies reviewed, the types of outcome measures used to assess clinical utility were highly variable with measures spanning each domain of the FT Model. Despite the presence of outcome measures in each of the efficacy domains, the largest proportion of outcome measures was assigned to the diagnostic accuracy domain. This pattern emerged despite our emphasis on clinical (and not diagnostic) utility-oriented search terms. This may reflect the field’s long-standing emphasis on laboratory performance as a core measure of utility. While strategies for measuring outcomes that extend beyond laboratory performance are now being articulated [7, 13,14,15, 19] and health technology assessment and reimbursement review bodies are beginning to require attention to an extended definition of utility [20, 21], it remains challenging to identify and measure meaningful health and non-health related outcomes attributable to genetic testing, to conduct robust studies in a manner that keeps pace with rapidly evolving technology platforms, and to incentivize this type of research when it is not uniformly required by regulatory or reimbursement bodies.
In addition, despite the broad range of outcomes identified, we were able to group the measures into a reduced set of thematic categories that map well to the FT Model. While narrow definitions of clinical utility may focus on a test’s ability to produce a diagnosis, broader definitions of clinical utility consider health and non-health related, familial and societal outcomes as proposed by the ACMG [9]. Outcome categories such as those identified in this review can be useful in consensus building efforts that aim to establish explicit and uniform strategies for measuring clinical utility, defined broadly. In addition to these outcomes, researchers may draw from the utility-related indices that have been identified in the literature [22] or other data elements that map onto specific utility-related categories. With respect to clinical indication for testing, oncology was most common (32.0%). This is likely the result of the growing role that genetic testing has in guiding cancer treatment selection and surveillance [23]. Consistent with the oncology emphasis in the literature, studies were also more likely to reflect on adult populations compared to pediatric rare disease populations where therapeutic options—and health outcomes research—are more limited. In addition, the majority of papers used an observational study design (88.7%), illustrating the relative absence of experimental approaches. In part, this may be related to the practical challenges associated with experimental study design in the context of rare disease and in part this may be related to the absence of regulatory requirements to conduct comparative effectiveness research for genome diagnostics [24, 25]. More robust study designs (i.e., prospective, comparative) and/or prioritizing genetic test evaluation research in a wider range of clinical settings (i.e., beyond cancer) may enable progress towards a richer evidentiary base for genetic testing, in turn guiding and substantiating efforts to adopt these technologies into health care systems. It is noteworthy that the diagnostic thinking efficacy domain (i.e., the test’s ability to help a clinician come to a diagnosis) had a limited presence in the reviewed literature; outcomes relevant to this domain were identified in only two studies We speculate the relative absence of this domain is tied to its close relationship with diagnostic accuracy efficacy; where diagnostic accuracy efficacy is achieved, diagnostic thinking efficacy may be assumed. This assumption may be unwarranted, however, because diagnostic accuracy measures do not capture the extent to which a test result helps a clinician come to a diagnosis and/or how the test results compare to a clinician’s pretest estimate of the probability of disease. Since diagnostic thinking refers to the subtle idea of how ‘helpful’ a variant is in contributing to a diagnosis or in planning subsequent steps in a diagnostic work up, it is challenging to measure. Moving downstream on the chain of evidence, diagnostic thinking efficacy may be implied where therapeutic efficacy has been demonstrated. Alternatively, evidence relevant to this efficacy domain may be addressed more commonly in literature that was deemed to be out of scope for this review. Future work to explore strategies for operationalizing measures of diagnostic thinking efficacy may be warranted.
This review also explored the ways in which genetic test purpose and clinical indication mapped onto the efficacy domains. In both cases, diagnostic accuracy outcomes were dominant. However, compared to the other test applications, the studies that evaluated tests with a diagnostic purpose had relatively low representation of downstream efficacy domains (i.e., beyond diagnostic accuracy). This suggests that value beyond the ability to establish a diagnosis was not well represented. For cancer, cardiac, and congenital anomalies indications, diagnostic accuracy outcomes were represented most frequently. However, the frequency of the other efficacy domains varied by clinical indication. This finding is consistent with the expectation that the clinical utility of genetic tests is context-dependent (i.e., test purpose and clinical indication). As such, the relevance of each efficacy domain may vary accordingly (Fig. 2).
Findings herein enable reflection on the suitability of the FT Model as an organizing framework for studies that evaluate genetic testing. First, the selected literature identified components of utility that may warrant inclusion in the FT Model, when applied to genetic testing. Additional measures included the impact of genetic testing on the estimated prevalence of disease, time to diagnosis, parent health outcomes, and genetic test utilization in different clinical settings. Additionally, the data extraction process revealed that clarifying the meaning of the term “therapeutic efficacy” may be warranted. While commonly used in epidemiology to describe the ability of a drug or a device to treat a disease, its use in the FT Model refers to a test’s impact on treatment planning. Taking this into consideration, a revised title for this domain (e.g., therapeutic thinking efficacy) may improve the clarity and applicability of the FT Model to the genetic testing context. Moreover, our data suggest that time to diagnosis be added to the diagnostic thinking efficacy domain and that parent or family-related outcomes warrant categorization. Arguably, parent and family-related outcomes sit at the boundary between patient outcome and societal efficacy domains as they represent a step beyond the index patient but a step shy of broad social considerations. Lastly, a permeable boundary between diagnostic accuracy and diagnostic thinking efficacy may be warranted to reflect the relatedness of these levels of utility. Table S4 presents suggested refinements to the FT Model.
This scoping review was limited by its relatively short time horizon (2015–2017), the inclusion of only English language studies, and a search strategy focused on only three domains of test efficacy. Although the catalog of measures produced from this review is not a comprehensive list of all possible outcome measures relevant to each domain of efficacy, the catalog may serve as a resource for the research and decision-making communities interested in advancing work in this area. Additionally, it should be acknowledged that the strategy used to identify articles for exclusion was limited by the EndNote X7-7.1 filters used and/or the reviewer’s interpretation of the information presented in the abstract or full-text article. For example, it is possible that case series that were not described as such in the article were included in the category of observational studies. Limitations notwithstanding, the results provide a rich description of the multi-dimensional nature of clinical utility and the current research activity that reflects its measurement [17].
In conclusion, findings herein serve to generate a better understanding of the range of strategies used to capture the concept of clinical utility in genetic medicine. Specifically, this review generated a thematically organized catalog of outcome measures that aim to capture the clinical utility of genetic testing: a catalog that can be used as a resource by researchers, clinicians, and decision makers in the field. Furthermore, having applied the FT Model as an organizational tool, thematic categories of clinical utility were also identified within the FT Model efficacy domains. As such, findings herein contribute to the community’s ongoing thinking about the ways in which the construct of clinical utility can be defined and operationalized. We also offer possible refinements to the FT Model based on our findings. These refinements to the framework and efforts to improve strategies for measuring value in genome medicine warrant further consideration.
References
Van ElCG, Cornel MC, Borry P, Hastings RJ, Fellmann F, Hodgson SV. et al. Whole-genome sequencing in health care. Eur J Hum Genet. 2013;21:580–4. https://doi.org/10.1038/ejhg.2013.46
Xue Y, Ankala A, Wilcox WR, Hegde MR. Solving the molecular diagnostic testing conundrum for Mendelian disorders in the era of next-generation sequencing: Single-gene, gene panel, or exome/genome sequencing. Genet Med. 2015;17:444–51.
Caulfield T, Evans J, McGuire A, McCabe C, Bubela T, Cook-Deegan R, et al. Reflections on the cost of “low-cost” whole genome sequencing: framing the health policy debate. PLoS Biol. 2013;11:7–12.
Nelson B. Ensuring quality in genomic medicine: amid the rise in complex laboratory-developed tests, regulatory officials are seeking the right balance on quality assurance. Cancer Cytopathol. 2014;122:855–6. http://doi.wiley.com/10.1002/cncy.21499
Sun F, Bruening W, Erinoff E, Schoelles KM. Addressing challenges in genetic test evaluation. addressing challenges genet test. Eval Eval Fram Assess Anal Validity. 2011. http://www.ncbi.nlm.nih.gov/pubmed/21834175
Grosse SD, Khoury MJ. What is the clinical utility of genetic testing? Genet Med. 2006;8:448–50. http://www.nature.com/doifinder/10.1097/01.gim.0000227935.26763.c6
Burke W. Genetic tests: clinical validity and clinical utility. Curr Protoc Hum Genet. 2014;81:9.15.1–8. http://www.ncbi.nlm.nih.gov/pubmed/24763995
National Academies of Sciences Engineering and Medicine. An evidence framework for genetic testing. National Academies of Sciences Engineering and Medicine; 2017.
Watson M. Clinical utility of genetic and genomic services: a position statement of the American College of Medical Genetics and Genomics. Genet Med. 2015;17:505–7.
Bossuyt PMM, Reitsma JB, Linnet K, Moons KGM. Beyond diagnostic accuracy: the clinical utility of diagnostic tests. Clin Chem. 2012;58:1636–43.
Garrison LP, Neumann PJ, Willke RJ, Basu A, Danzon PM, Doshi JA. et al. A health economics approach to US value assessment frameworks—summary and recommendations of the ISPOR Special Task Force Report [7]. Value Heal. 2018;21:161–5. https://doi.org/10.1016/j.jval.2017.12.009
Pitini E, Vito C De, Marzuillo C, Andrea ED, Rosso A, Federici A, et al. How is genetic testing evaluated? A systematic review of the literature. Eur J Hum Genet. 2017;605–15. https://doi.org/10.1038/s41431-018-0095-5
Teutsch SM, Bradley LA, Palomaki GE, Haddow JE, Piper M, Calonge N, et al. The evaluation of genomic applications in practice and prevention (EGAPP) initiative: methods of the EGAPP working group. Hum Genome Epidemiol Build Evid Using Genet Inf Improv Heal Prev Dis Second Ed. 2010;11:3–14.
Agency for Healthcare Research and Quality. Methods Guide for Medical Test reviews. Agency for Healthcare Research and Quality; 2012.
Ferrante Di Ruffano L, Hyde CJ, McCaffery KJ, Bossuyt PMM, Deeks JJ. Assessing the value of diagnostic tests: a framework for designing and evaluating trials. BMJ. 2012;344:1–9.
Fryback DG, Thornbury JR. The efficacy of diagnostic imaging. Med Decis Mak. 1991;11:88–94.
Arksey H, O’Malley L. Scoping studies: towards a methodological framework. Int J Soc Res Methodol Theory Pr. 2005;8:19–32.
WHO. Statistical classification of diseases and related health problems 10th revision. World Health Organization; 2016. http://www.who.int/classifications/icd/en/
Botkin JR, Teutsch SM, Kaye CI, Hayes M, Haddow JE, Bradley LA, et al. Outcomes of interest in evidence-based evaluations of genetic tests. Genet Med. 2010;12:228–35. http://www.nature.com/doifinder/10.1097/GIM.0b013e3181cdde04
European Commission. Regulatory framework: the new Regulations on medical devices. European Commision; 2018. http://ec.europa.eu/growth/sectors/medical-devices/regulatory-framework/
Ontario Health Technology Advisory Committee (OHTAC). Decision determinants guidance document: the Ontario Health technology advisory committee (OHTAC) decision-making process for the development of evidence-based recommendations. Ontario Health Technology Advisory Committee; 2010. http://www.health.gov.on.ca/english/providers/program/mas/pub/guide_decision.pdf
Hayeems RZ, Luca S, Ungar WJ, Bhatt A, Chad L, Pullenayegum E. et al. The development of the Clinician-reported Genetic testing Utility InDEx (C-GUIDE): a novel strategy for measuring the clinical utility of genetic testing. Genet Med. 2020;22:95–101. https://doi.org/10.1038/s41436-019-0620-0
NIH National Cancer Institute. Cancer genomics research. 2018. https://www.cancer.gov/research/areas/genomics
Mccarthy JJ, Mcleod HL, Ginsburg GS. Genomic medicine: a decade of successes, challenges, and opportunities. Sci Transl Med. 2013;5:189sr4. https://doi.org/10.1126/scitranslmed.3005785
Calonge N, Klein RD, Berg AO, Berg JS, Armstrong K, Botkin J, et al. The EGAPP initiative: lessons learned. Genet Med. 2014;16:217–24.
Acknowledgements
This work was funded by the Canadian Institutes of Health Research Project Grant (PJT-152880).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Walcott, S.E., Miller, F.A., Dunsmore, K. et al. Measuring clinical utility in the context of genetic testing: a scoping review. Eur J Hum Genet 29, 378–386 (2021). https://doi.org/10.1038/s41431-020-00744-2
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41431-020-00744-2
This article is cited by
-
Applying the Clinician-reported Genetic testing Utility InDEx (C-GUIDE) to genome sequencing: further evidence of validity
European Journal of Human Genetics (2022)
-
2021 at European Journal of Human Genetics: the year in review
European Journal of Human Genetics (2022)
-
Genomic health data generation in the UK: a 360 view
European Journal of Human Genetics (2022)