Abstract
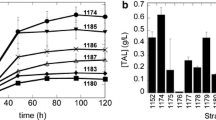
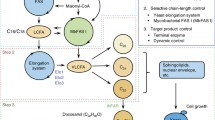
Adipic Acid (AA) is a valued platform chemical compound, which can be used as a precursor of nylon-6,6. Due to the generation of an enormous amount of nitric oxide metabolites and the growing depletion of oil resources as a result of AA production from a mixture of cyclohexanol and cyclohexanone, the microbial methods for synthesizing AA have attracted significant attention. Of the several AA-producing pathways, the reverse adipate degradation pathway in Thermobifida fusca (Tfu RADP) is reported to be the most efficient, which has been confirmed in Escherichia coli. In this study, the heterologous Tfu RADP was constructed for producing AA in S. cerevisiae by co-expressing genes of Tfu_0875, Tfu_2399, Tfu_0067, Tfu_1647, Tfu_2576, and Tfu_2576. The AA titer combined with biomass, cofactors and other by-products was all determined after fermentation. During batch fermentation in a shake flask, the maximum AA titer was 3.83 mg/L, while the titer increased to 10.09 mg/L during fed-batch fermentation in a 5-L bioreactor after fermentation modification.
Similar content being viewed by others
References
Alini, S., Basile, F., Blasioli, S., Rinaldi, C., and Vaccari, A. 2007. Development of new catalysts for N2O-decomposition from adipic acid plant. Appl. Catal. B 70, 323–329.
Armstrong, K.A., Som, T., Volkert, F.C., Rose, A., and Broach, J.R. 1989. Propagation and expression of genes in yeast using 2-micron circle vectors. Biotechnology 13, 165–192.
Averesch, N.J.H., Martínez, V.S., Nielsen, L.K., and Krömer, J.O. 2018. Towards synthetic biology strategies for adipic acid production — an in-silico tool for combined thermodynamics and stoichiometric analysis of metabolic networks. ACS Synth. Biol. 7, 490–509.
Bart, J.C.J. and Cavallaro, S. 2015. Transiting from adipic acid to bio-adipic acid Part II. Biosynthetic pathways. Ind. Eng. Chem. Res. 54, 567–576.
Beardslee, T. and Picataggio, S. 2012. Bio-based adipic acid from renewable oils. Lipid Tech. 24, 223–225.
Bernofsky, C. and Swan, M. 1973. An improved cycling assay for nicotinamide adenine dinucleotide. Anal. Biochem. 53, 452–458.
Bozell, J.J. and Petersen, G.R. 2010. Technology development for the production of biobased products from biorefinery carbohydrates — the US Department of Energy’s “Top 10” revisited. Green Chem. 12, 539–554.
Brindle, N.P., Zammit, V.A., and Pogson, C.I. 1985. Regulation of carnitine palmitoyltransferase activity by malonyl-CoA in mitochondria from sheep liver, a tissue with a low capacity for fatty acid synthesis. Biochem. J. 232, 177–182.
Burgard, A., Pharkya, P., and Osterhout, R. 2010. Microorganisms for the production of adipic acid and other compounds. US Patent No.7,799,545 B2. Genomatica, Inc., San Diego, California, USA.
Canelas, A.B., van Gulik, W.M., and Heijnen, J.J. 2008. Determination of the cytosolic free NAD/NADH ratio in Saccharomyces cerevisiae under steady-state and highly dynamic conditions. Biotechnol. Bioeng. 100, 734–743.
Chaustova, L., Miliukienė, V., and Zimkus, A. 2010. Cell de-energization prevents plasmid transformation of yeast Saccharomyces cerevisiae: evidence for the requirement of ATP. Cent. Eur. J. Biol. 5, 78–82.
Cheong, S., Clomburg, J.M., and Gonzalez, R. 2016. Energy- and carbon-efficient synthesis of functionalized small molecules in bacteria using non-decarboxylative Claisen condensation reactions. Nat. Biotechnol. 34, 556–561.
Clomburg J.M., Blankschien, M.D., Vick, J.E., Chou, A., Kim, S., and Gonzalez, R. 2015. Integrated engineering of β-oxidation reversal and ω-oxidation pathways for the synthesis of medium chain ω-functionalized carboxylic acids. Metab. Eng. 28, 202–212.
Deng Y. and Fong, S.S. 2011. Metabolic engineering of Thermobifida fusca for direct aerobic bioconversion of untreated lignocellulosic biomass to 1-propanol. Metab. Eng. 13, 570–577.
Deng, Y. and Mao, Y. 2015. Production of adipic acid by the native-occurring pathway in Thermobifida fusca B6. J. Appl. Microbiol. 119, 1057–1063.
Elgersma, Y., van Roermund, C.W., Wanders, R.J., and Tabak, H.F. 1995. Peroxisomal and mitochondrial carnitine acetyltransferases of Saccharomyces cerevisiae are encoded by a single gene. EMBO J. 14, 3472–3479.
Flagfeldt, D.B., Siewers, V., Huang, L., and Nielsen, J. 2010. Characterization of chromosomal integration sites for heterologous gene expression in Saccharomyces cerevisiae. Yeast 26, 545–551.
Frenkel, E.P. and Kitchens, R.L. 1981. Acetyl-CoA synthetase from bakers’ yeast (Saccharomyces cerevisiae): EC 6.2.1.1 acetate:CoA ligase (AMP-forming). In Lowenstein, J.M. (ed.), Methods in Enzymology, vol. 71, pp. 317–324. Academic Press, Cambridge, Massachusatts, USA.
Ganzhorn, A.J., Green, D.W., Hershey, A.D., Gould, R.M., and Plapp, B.V. 1987. Kinetic characterization of yeast alcohol dehydrogenases. Amino acid residue 294 and substrate specificity. J. Biol. Chem. 262, 3754–3761.
Gibson, D.G. 2014. Programming biological operating systems: genome design, assembly and activation. Nat. Methods 11, 521–526.
Gibson, D.G., Young, L., Chuang, R.Y., Venter, J.C., Hutchison, C.A., and Smith, H.O. 2009. Enzymatic assembly of DNA molecules up to several hundred kilobases. Nat. Methods 6, 343–345.
Gietz, R.D. and Woods, R.A. 2006. Yeast transformation by the LiAc/SS carrier DNA/PEG method. In Xiao, W. (ed.), Yeast Protocol. Methods in Molecular Biology, vol. 313. pp. 107–120. Humana Press, Totowa, New Jersey, USA.
Gunukula, S. and Anex, R.P. 2017. Techno-economic analysis of multiple bio-based routes to adipic acid. Biofuels Bioprod. Bioref. 11, 897–907.
Heux, S., Cachon, R., and Dequin, S. 2006. Cofactor engineering in Saccharomyces cerevisiae: expression of a H2O-forming NADH oxidase and impact on redox metabolism. Metab. Eng. 8, 303–314.
Huang, D., Yang, K., Liu, J., Xu, Y., Wang, Y., Wang, R., Liu, B., and Feng, L. 2017. Metabolic engineering of Escherichia coli for the production of 2′-fucosyllactose and 3-fucosyllactose through modular pathway enhancement. Metab. Eng. 41, 23–38.
Jou, W.S., Chen, K.N., Chao, D.Y., Lin, C.Y., and Yeh, J.T. 2001. Flame retardant and dielectric properties of glass fibre reinforced nylon-66 filled with red phosphorous. Polym. Degrad. Stab. 74, 239–245.
Koebmann, B.J., Westerhoff, H.V., Snoep, J.L., Dan, N., and Jensen, P.R. 2002a. The glycolytic flux in Escherichia coli is controlled by the demand for ATP. J. Bacteriol. 184, 3909–3916.
Koebmann, B.J., Westerhoff, H.V., Snoep, J.L., Solem, C., Pedersen, M.B., Nilsson, D., Michelsen, O., and Jensen, P.R. 2002b. The extent to which ATP demand controls the glycolytic flux depends strongly on the organism and conditions for growth. Mol. Biol. Rep. 29, 41–45.
Kruyer, N.S. and Peralta-Yahya, P. 2017. Metabolic engineering strategies to bio-adipic acid production. Curr. Opin. Biotechnol. 45, 136–143.
Leavitt, J.M., Wagner, J.M., Tu, C.C., Tong, A., Liu, Y., and Alper, H.S. 2017. Biosensor-enabled directed evolution to improve muconic acid production in Saccharomyces cerevisiae. Biotechnol. J. 12, 1600687.
Lee, F.W. and Da Silva, N.A. 1997. Improved efficiency and stability of multiple cloned gene insertions at the delta sequences of Saccharomyces cerevisiae. Appl. Microbiol. Biotechnol. 48, 339–345.
Lee, K.M. and Da Silva, N.A. 2005. Evaluation of the Saccharomyces cerevisiae ADH2 promoter for protein synthesis. Yeast 22, 431–440.
Lin, S.J., Ford, E., Haigis, M., Liszt, G., and Guarente, L. 2004. Calorie restriction extends yeast life span by lowering the level of NADH. Genes Dev. 18, 12–16.
Liu, J., Killilea, D.W., and Ames, B.N. 2002. Age-associated mitochondrial oxidative decay: Improvement of carnitine acetyltransferase substrate-binding affinity and activity in brain by feeding old rats acetyl-L-carnitine and/or R-α-lipoic acid. Proc. Natl. Acad. Sci. USA 99, 1876–1881.
Livak, K.J. and Schmittgen, T.D. 2001. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔct method. Methods 25, 402–408.
Lutstorf, U. and Megnet, R. 1968. Multiple forms of alcohol dehydrogenase in Saccharomyces cerevisiae: I. Physiological control of ADH-2 and properties of ADH-2 and ADH-4. Arch. Biochem. Biophys. 126, 933–944.
Meaden, P.G., Dickinson, F.M., Mifsud, A., Tessier, W., Westwater, J., Bussey, H., and Midgley, M. 1997. The ALD6 gene of Saccharomyces cerevisiae encodes a cytosolic, Mg2+-activated acetaldehyde dehydrogenase. Yeast 13, 1319–1327.
Mizuno, A., Tabei, H., and Iwahuti, M. 2006. Characterization of low-acetic-acid-producing yeast isolated from 2-deoxyglucose-resistant mutants and its application to high-gravity brewing. J. Biosci. Bioeng. 101, 31–37.
Nakamura, S., Takasaki, H., Kobayashi, K., and Kato, A. 1993. Hyperglycosylation of hen egg white lysozyme in yeast. J. Biol. Chem. 268, 12706–12712.
Parekh, R.N., Shaw, M.R., and Wittrup, K.D. 1996. An integrating vector for tunable, high copy, stable integration into the dispersed Ty δ sites of Saccharomyces cerevisiae. Biotechnol. Prog. 12, 16–21.
Przybyla-Zawislak, B., Dennis, R.A., Zakharkin, S.O., and McCammon, M.T. 1998. Genes of succinyl-CoA ligase from Saccharomyces cerevisiae. Eur. J. Biochem. 258, 736–743.
Raj, K., Partow, S., Correia, K., Khusnutdinova, A.N., Yakunin, A.F., and Mahadevan, R. 2018. Biocatalytic production of adipic acid from glucose using engineered Saccharomyces cerevisiae. Metab. Eng. Commun. 6, 28–32.
Romanos, M.A., Scorer, C.A., and Clare, J.J. 1992. Foreign gene expression in yeast: a review. Yeast 8, 423–488.
San, K.Y., Bennett, G.N., Berrios-Rivera, S.J., Vadali, R.V., Yang, Y.T., Horton, E., Rudolph, F.B., Sariyar, B., and Blackwood, K. 2002. Metabolic engineering through cofactor manipulation and its effects on metabolic flux redistribution in Escherichia coli. Metab. Eng. 4, 182–192.
Skoog, E., Shin, J.H., Saez-Jimenez, V., Mapelli, V., and Olsson, L. 2018. Biobased adipic acid — The challenge of developing the production host. Biotechnol. Adv. 36, 2248–2263.
Smith, M.G., Des Etages, S.G., and Snyder, M. 2004. Microbial synergy via an ethanol-triggered pathway. Mol. Cell. Biol. 24, 3874–3884.
Suástegui, M., Ng, C.Y., Chowdhury, A., Sun, W., Cao, M., House, E., Maranas, C.D., and Shao, Z. 2017. Multilevel engineering of the upstream module of aromatic amino acid biosynthesis in Saccharomyces cerevisiae for high production of polymer and drug precursors. Metab. Eng. 42, 134–144.
Yu, J.L., Xia, X.X., Zhong, J.J., and Qian, Z.G. 2014. Direct biosynthesis of adipic acid from a synthetic pathway in recombinant Escherichia coli. Biotechnol. Bioeng. 111, 2580–2586.
Zhao, M., Huang, D., Zhang, X., Koffas, M.A.G., Zhou, J., and Deng, Y. 2018. Metabolic engineering of Escherichia coli for producing adipic acid through the reverse adipate-degradation pathway. Metab. Eng. 47, 254–262.
Acknowledgments
This work was funded by grants from the National Key R&D Program of China (2019YFA0905502), the National Natural Science Foundation of China (21877053, 31601564), the Natural Science Foundation of Jiangsu Province (BK20181345), the Fundamental Research Funds for the Central Universities (JUSRP51705A), the Open Foundation of Jiangsu Key Laboratory of Industrial Biotechnology (KLIB-KF201807), and the Postgraduate Research & Practice Innovation Program of Jiangsu Provence (KYCX17_1424).
Author information
Authors and Affiliations
Corresponding authors
Additional information
Conflict of Interest
All authors declare that they have no conflicts of interest with this manuscript.
Supplemental material for this article may be found at http://www.springerlink.com/content/120956.
Electronic supplementary material
Rights and permissions
About this article
Cite this article
Zhang, X., Liu, Y., Wang, J. et al. Biosynthesis of adipic acid in metabolically engineered Saccharomyces cerevisiae. J Microbiol. 58, 1065–1075 (2020). https://doi.org/10.1007/s12275-020-0261-7
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12275-020-0261-7