Abstract
The ongoing pandemic caused by severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) has affected millions of people worldwide and with notable heterogeneity in its clinical presentation. Probability of contracting this highly contagious infection is similar across age groups but disease severity and fatality among aged patients with or without comorbidities are reportedly higher. Previous studies suggest that age associated transcriptional changes in lung and immune system results in a proinflammatory state and increased susceptibility to infectious lung diseases. Similarly, SARS-CoV-2 infection could augment ageing-related gene expression alterations resulting in severe outcomes in elderly patients. To identify genes that can potentially increase covid-19 disease severity in ageing people, we compared age associated gene expression changes with disease-associated expression changes in lung/BALF and whole blood obtained from publicly available data. We observed (i) a significant overlap of gene expression profiles of patients’ BALF and blood with lung and blood of the healthy group, respectively; (ii) a more pronounced overlap in blood compared to lung; and (iii) a similar overlap between host genes interacting with SARS-CoV-2 and ageing blood transcriptome. Pathway enrichment analysis of overlapping gene sets suggest that infection alters expression of genes already dysregulated in the elderly, which together may lead to poor prognosis. eQTLs in these genes may also confer poor outcome in young patients worsening with age and comorbidities. Further, the pronounced overlap observed in blood may explain clinical symptoms including blood clots, strokes, heart attack, multi-organ failure etc. in severe cases. This model based on a limited patient dataset seems robust and holds promise for testing larger tissue specific datasets from patients with varied severity and across populations.
Similar content being viewed by others
Introduction
Since December 2019, severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), that causes COVID-19 in humans (Siordia 2020) has spread rapidly across the world and is classified as a global pandemic by the World Health Organization (coronavirus (COVID-19) events as they happen; https://www.who.int/emergencies/diseases/novel-coronavirus-2019/events-as-they-happen). As of 2 July 2020, there have been 10.6 million confirmed COVID-19 cases with 519,766 deaths reported worldwide (https://ourworldindata.org), with continuing trend of sharp rise in both these categories in many countries/regions. The disease is highly heterogenous in its clinical presentation with most common symptoms being fever, cough, shortness of breath and fatigue. In addition, myalgia, neurological symptoms, ischaemic and haemorrhagic strokes, muscle injury and gastrointestinal symptoms are also reported in a subset of patients (Harapan et al. 2020). Several clinical trials are ongoing but as of date, no drugs or other therapeutics have been approved by the U.S. Food and Drug Administration (FDA) to prevent or treat COVID-19 (Sanders et al. 2020) and thus clinical management includes infection prevention and supportive care (Sanders et al. 2020). Efforts to combat COVID-19 are severely hampered by grossly inadequate knowledge of several important aspects of the illness ranging from pathogen biology to host response, disease biology, target tissues and consequently treatment options. Therefore, there is an urgent need for a deeper understanding of the host–pathogen interaction biology of SARS-CoV-2, which in turn may offer important insights into general/personalized treatment strategies and management of the disease as well as development of new therapies.
Probability of contracting this highly contagious infection has been reported to be similar across age groups but severe clinical manifestations and increased mortality has been reported in elderly patients (Cohen et al. 2020; Hauser et al. 2020; Meyerowitz-Katz and Merone 2020; Wu et al. 2020). These observations suggest that there may be a comparatively stronger association between age and poor prognosis of COVID-19, but this may well be multifactorial. Therefore, the uncovering mechanism(s) underlying poor prognosis to SARS-CoV-2 infection among the affected elderly might be insightful for effective patient management and treatment. Several reports suggest that with the ageing, elderly population becomes more susceptible to various infectious disorders (Meyer 2001; Gavazzi and Krause 2002; López-Otín et al. 2003; Meiners et al. 2015). With ageing, transcriptional dysregulation occurs in genes involved in cellular oxidant/antioxidant systems, proinflammatory mediators (C-reactive protein, tumour necrosis factor α (TNF-α), interleukins (IL) 6, 1β), and cell regenerations that might overlap with viral mediated dysregulation (Franceschi and Campisi 2014; Meiners et al. 2015; de Almeida et al. 2020; Fulop et al. 2018). Similarly in severe COVID-19 patients, a ‘cytokine storm’ comprised of TNF-α, IL 6, 1β, 8, 12, interferon-gamma inducible protein (IP10), macrophage inflammatory protein 1A (MIP1A), and monocyte chemoattractant protein 1 (MCP1) (Cascella et al. 2020) and hypercoagulable state with increased risk of venous thromboembolism (Cevik et al. 2020) has been observed.
Based on this limited understanding, we hypothesize that SARS-CoV-2 mediated transcriptional alterations may overlap with age mediated expression changes. Thus, expression of genes that changes during ageing, might get further augmented on SARS-CoV-2 infection, leading to severe outcome in elderly patients. We attempted to explore this possibility by performing comparative transcriptomics using available data from two target tissues, namely lung and blood in healthy ageing group and COVID-19 patients. We also compared transcriptomic profile of ageing lung and blood with host genes interacting with SARS-CoV-2 protein. We observed a significant overlap between gene expression profile in both lung and blood of healthy ageing group and COVID-19 patients; which was much more pronounced in blood. Further, there was a significant overlap between host genes interacting SARS-CoV-2 proteins in ageing blood but not in lungs. These observations support previous reports that SARS-COV-2, primarily affects the respiratory system but its effects may manifest in blood leading to multiorgan failure in severe cases of COVID-19 (Cascella et al. 2020; Varga et al. 2020).
Materials and methods
Study design
Identification of candidate genes
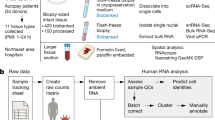
To identify the genes that determine poor prognosis in the elderly COVID-19 patients, we performed a comparative analysis of gene expression data as collected from bronchoalveolar-lavage fluid (BALF)/lung/blood from COVID-19 patients and healthy ageing group; and data of host genes interacting with SARS-CoV-2 proteins with transcription profile of ageing lung and blood (resources are mentioned below). This methodology was adopted from previous studies (Dugo et al. 2016; Elko et al. 2019). Overlaps between the following groups were documented: (i) patients’ BALF with healthy ageing lung; (ii) patients’ PBMCs with healthy ageing blood; (iii) host genes interacting with viral proteins with healthy ageing lung and blood.
Statistical analysis
Statistical significance of these overlaps, if any, were tested using hypergeometric test (http://nemates.org/MA/progs/overlap_stats_prog.html). Basic equation to find the probability of finding an overlap of genes using the above-mentioned program is provided in supplementary text.
Pathway analysis
Pathway enrichment of the significantly overlapping genes identified above was done using EnrichR (Chen et al. 2013; Kuleshov et al. 2016) which is an integrative web-based and mobile software application that currently includes 180,184 annotated gene sets from 102 gene set libraries and various interactive visualization approaches to display enrichment results using the JavaScrit library, Data Driven Documents.
eQTL analysis
eQTL variants of genes from significantly overlapping gene-sets for respective tissues (lung and blood) were obtained from GTEx v. 8 (https://gtexportal.org). With a view to obtain a universally applicable biomarker, a comparable minor allele frequency of the markers would be ideal and such a suitability was tested using FST or fixation index. This was done using 1000 genome phase 1 data with the help of an online tool SPSmart (http://spsmart.cesga.es) with FST (i) 0 to 0.05 representing low; (ii) 0.05 to 0.15, moderate; (iii) 0.15 to 0.25, high; and (iv) > 0.25, very high, genetic differentiation.
Identification of druggable targets
Finally, these genes identified above were screened for their druggability with FDA approved drugs using DGIdb website (http://dgidb.org).
Resources
Age associated genes
(i) Differentially expressed genes (DEGs) in lung and blood tissue identified in a recently published study using RNA-Seq based transcriptome profiles from human donors of various ages from GTEx (Yang et al. 2015) were enlisted. (ii) DEGs from two stage transcriptomic study performed in blood, based on meta-analysis data from six different studies (n = 7074 samples) in the discovery phase and 7909 additional whole-blood samples in the replication phase (Peters et al. 2015) were obtained.
These two datasets were merged for blood transcriptomics and all the protein coding genes (and not any small RNAs such as miRNA, lncRNA etc.) that were reported in either study were considered. All genes showing a different direction of expression change were removed from the analysis. A total of 2877 and 2283 protein coding genes were found to be upregulated and downregulated, respectively in blood of healthy ageing group. Similarly, a total of 363 and 592 protein coding genes were found to be upregulated and downregulated, respectively in ageing lung. These genes have been subsequently referred to as ‘age-associated DEGs’.
SARS-CoV-2 associated genes
Henceforth the DEGs in COVID-19 positive patients are referred to as ‘SARS-CoV-2-associated DEGs’ and were obtained from two recent studies: (i) DEGs in BALFs identified comparing laboratory-confirmed COVID-19 patients (SARS2) (n = 8, median age 50.5 years) with healthy controls without known respiratory diseases (healthy) (n = 20) (Zhou et al. 2020). (ii) DEGs in PBMCs and BALF identified by comparing three COVID-19 patients (median age 37 years) and three healthy donors (Xiong et al. 2020).
BALF transcriptomics from these two datasets were merged and all the protein coding genes (and not any small RNAs such miRNA, lncRNA etc.) that are reported in either study were considered. All genes showing a different direction of expression change were removed from the analysis.
Host genes interacting with SARS-CoV-2 proteins
Host genes that were found to be interacting with SARS-COV-2 viral proteins henceforth referred as ‘SARS-CoV2-interacting genes’ and were collected from recent studies mentioned below. (i) Three hundred and thirty-two high-confidence SARS-CoV-2-human protein–protein interactions (PPIs) were obtained by analysing the data generated by expressing 26 of the 29 SARS-CoV-2 proteins in HEK293 cells in a recent study (Gordon et al. 2020). (ii) Computation and literature based interactome data that were generated using available sequences for viral protein candidates (such as wS, wORF3a, wE, wM, wORF6, wORF7a, wORF7b, wORF8, wN and wORF10) in a recent study (Srinivasan et al. 2020).
Results
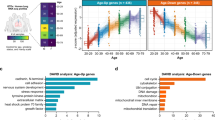
Test of hypothesis (figure 1) by a comparative analysis of DEGs in the different sample sets described under study design (figure 2) revealed significant overlaps between them (table 1; table 1 in electronic supplementary material at http://www.ac.in/jgenet/), which are briefly presented below (table 1; table 1 in electronic supplementary material), are briefly presented below. The most notable findings include (i) significant overlap (P< 1.4E-04) between the upregulated SARS-CoV-2 associated DEGs in patients’ BALF and upregulated age associated DEGs in healthy ageing. (ii) Significant overlap (P< 6.53E-07) between the upregulated SARS-CoV-2 associated DEGs in patients’ PBMCs and upregulated age associated DEGs in healthy ageing blood. (iii) Nominally significant overlap (P< 0.03) between the downregulated SARS-CoV-2 associated DEGs in patients’ PBMCs and downregulated age associated DEGs in healthy ageing blood. (iv) Significant overlap between the SARS-CoV-2 interacting genes and up (P< 0.002) / down (P< 1.04E-06) regulated age associated DEGs in healthy ageing blood (table 1; table 1 in electronic supplementary material).
Pathway enrichment
As the number of genes in each gene-set was very small to identify the enriched pathways, if any, all the genes from significant gene-sets and showing change in same direction were considered together for pathway enrichment analysis. Cytokine genes that are frequently found to be upregulated in patients and which overlapped with healthy ageing expression profiles were also included. Six of the 21 upregulated cytokine genes in patients, overlapped with ageing related genes in the blood but none with ageing lung. Upregulated genes in the healthy ageing group that overlap with SARS-CoV-2 associated genes and SARS-CoV-2 interacting genes were seen to be enriched in a range of signalling pathways, including p53, chemokine and cytokine mediated inflammation, EGF receptor, TGF-beta, AGE-RAGE, Toll-like receptor mediated, NF-kappa B, VEGFA-VEGFR2 and genes involved in ROS in triggering vascular inflammation, oxidoreductive damage, T cell polarization, lung fibrosis, chronic obstructive pulmonary disorder (COPD), and local acute inflammatory response (representative pictures at figure 3; table 2 in electronic supplementary material). Downregulated genes in the healthy ageing group that overlap SARS-CoV-2 associated and SARS-CoV-2 interacting genes were seen to be enriched in pathways such as PI3K-Akt-mTOR-signalling, membrane trafficking, HIV and influenza RNA transport, ISG15 antiviral mechanism, cellular export machinery that interacts with NEP/NS2 (representative pictures at figure 4; table 3 in electronic supplementary material).
eQTL analysis
eQTL variants in genes from significantly overlapping gene-sets from the respective tissues namely lung and blood were identified (table 2; table 4 in electronic supplementary material). FST test was performed to identify the variants with low genetic differentiation among different population. A large number of eQTL variants with FST< 0.05 were found in each group (table 2; table 4 in electronic supplementary material). Further analysis of these may enable identification of variant(s) which could be used as a biomarker(s).
Identification of druggable gene targets
It may be mentioned that SARS-CoV-2 interacting genes were excluded from this analysis since they have already been reported previously (Gordon et al. 2020). Efforts to identify novel druggable targets for FDA approved drugs from among 259 genes from significantly overlapping gene sets between different groups shown above (table 1), yielded a total of 48 druggable genes mostly from the immune system related pathways. A total of 205 FDA approved drugs could potentially target them (table 5 in electronic supplementary material).
Discussion
COVID-19, the recent pandemic has affected individuals of all age groups as well as all ethnicities. However, poor prognosis has been witnessed in the elderly effected group with or without comorbidities (Hauser et al. 2020; Cohen et al. 2020; Meyerowitz-Katz and Merone 2020; Wu et al. 2020). The overall case fatality rate (CFR) for cases with age 70 to 79 years is 8.0% and for cases with age 80 years and above it is 14.8%; which is strikingly higher compared to 0.4% in cases below 50 years of age, based on a study that included 72314 cases from China (Wu and McGoogan 2020). It is well known that with ageing there is a notable increase in circulating proinflammatory cytokines even in the absence of an immunological threat and also a reduction in proteins that maintain homeostasis of the immune system, thus entailing a greater risk for many diseases (cancer, cardiovascular and neurodegenerative disorders, COPD, lung cancer, interstitial lung disease etc.) (Panda et al. 2009; López-Otín et al. 2013; Meiners et al. 2015; Angelidis et al. 2019). Thus, we reasoned that comparing naturally occurring, age-dependent transcriptional changes with those observed in COVID19 patients and/or known SARS-COV-2 interacting proteins may provide insights into the age-associated poor prognosis in COVID-19.
Three noteworthy findings emerged from our study: (i) a significant overlap was witnessed between DEGs in patients’ BALF/blood and in ageing lung and blood; (ii) this overlap was more pronounced in blood compared to lung; and (iii) a similar overlap between SARS-CoV-2 interacting proteins and DEGs in ageing blood but not in lung (table 1) and warrant discussion. Pathway enrichment analysis of the overlapped gene sets suggest that genes involved in pathways such as proinflammatory, apoptotic, T cell polarization, viral replication suppression (figures 3 & 4; tables 2 & 3 in electronic supplementary material) remain dysregulated in elderly patients. Upon SARS-CoV-2 infection, expression of these genes gets further dysregulated resulting in poor prognosis in case of elderly patients.
As for the second novel observation, the higher extent of overlap between SARS-CoV-2 associated / SARS-CoV-2 interacting genes and DEGs in blood among the heathy ageing group, may explain the range of clinical symptoms including high prevalence of blood clots, strokes and heart attack as well as multi-organ failure in a subset of severe patients reported in this disorder (Harapan et al. 2020; Cevik et al. 2020). Our observations are corroborated by a recent report of SARS-CoV-2 mediated damage to endothelial cells lining the blood vessels probably leading to blood clotting, strokes and heart attacks (Varga et al. 2020). We observed that proinflammatory genes such as IFNG, CCL4, CCR9, BCL2, TIMP1, TNF, C2, CCR2, VEGFA (table 1 in electronic supplementary material) remain upregulated in aged individuals which might cause the ‘cytokine storm’ observed in severe patients. Further, higher expression of these genes may lead to over activation of T cell polarization and TGF-beta pathways in affected elderly patients (figure 3), which can cause functional exhaustion of T cells (Swain et al. 2012; Kahan et al. 2015) that has been reported in elderly patients (Diao et al. 2020). Of note, one of these genes, namely CCR9 was found to be significantly (P = 1.15 × 10−10) associated with COVID-19 in a recent genomewide association study that included 835 patients and 1255 control participants from Italy and 775 patients and 950 control subjects from Spain (Ellinghaus et al. 2020), lending support to our findings.
Taken together these observations, eQTLs in the genes identified in our study (table 2; table 4 in electronic supplementary material) may also confer poor prognosis in young patients but this may worsen with age and comorbidities. However, their utility as prognostic biomarkers across different populations may be assessed only when more data of patients become available and these genes are characterized as COVID-19 patients. On the other hand, variants with FST > 0.05 may be tested for correlation with population specific severity. Additionally, identification of genes involved in influenza and HIV infection in our study (figures 3&4; tables 2&3 in electronic supplementary material) may be of considerable relevance for drug repurposing for treatment of COVID-19 patients.
In summary, our analysis (i) identified probable candidate genes for poor prognosis among the affected elderly group; (ii) identified potential druggable targets as well as FDA approved drugs opening the possibility of drug repurposing; and (iii) seems to provide early explanation for manifestation of blood related symptoms and probably multiorgan damage. Finally, although these leads are preliminary based on a very limited patient dataset, the model holds promise to be tested as and when more tissue specific patient derived data from different age groups and/or from different populations become available. Our study is limited by nonavailability of large tissue specific transcriptomic datasets of COVID-19 patients across age groups, and with varying severity and from different populations to validate the hypothesis.
References
Angelidis I., Simon L. M., Fernandez I. E., Strunz M., Mayr C. H., Greiffo F. R. et al. 2019 An atlas of the aging lung mapped by single cell transcriptomics and deep tissue proteomics. Nat. Commun. 10. https://doi.org/10.1038/s41467-019-08831-9.
Cascella M., Rajnik M., Cuomo A., Dulebohn S. C., Di Napoli R. 2020 Features, evaluation and treatment of coronavirus (COVID-19). StatPearls; http://www.ncbi.nlm.nih.gov/pubmed/32150360.
Cevik M., Bamford C. and Ho A. 2020 COVID-19 pandemic - A focused review for clinicians. Clin. Microbiol. Infec. 26, 842–847.
Chen E. Y., Tan C. M., Kou Y., Duan Q., Wang Z., Meirelles G. V. et al. A. 2013 Enrichr: interactive and collaborative HTML5 gene list enrichment analysis tool. BMC Bioinformatics 14. https://doi.org/10.1186/1471-2105-14-128.
Cohen J. F., Korevaar D. A., Matczak S., Brice J., Chalumeau M. and Toubiana J. 2020 COVID-19-related mortality by age groups in Europe: A meta-analysis. MedRxiv, https://doi.org/10.1101/2020.04.11.20061721.
de Almeida A. J. P. O., de Almeida Rezende M. S., Dantas S. H., de Lima Silva S., de Oliveira J. C. P. L. et al. 2020 Unveiling the role of inflammation and oxidative stress on age-related cardiovascular diseases. Oxid. Med. Cell. Longev. 2020, 1–20.
Diao B., Wang C., Tan Y., Chen X., Liu Y., Ning L. et al. 2020. Reduction and functional exhaustion of T cells in patients with coronavirus disease 2019 (COVID-19). Front. Immunol. 11, 827.
Dugo M., Cotroneo C. E., Lavoie-Charland E., Incarbone M., Santambrogio L., Rosso L. et al. 2016 Human lung tissue transcriptome: influence of sex and age. PLoS One 11, e0167460.
Elko E. A., Mahoney J. M., Vacek P., van der Vliet A., Anathy V., van der Velden L. J. L. et al. 2019 Age-dependent dysregulation of redox genes may contribute to fibrotic pulmonary disease susceptibility. Free Radic. Biol. Med. 141, 438–446.
Ellinghaus D., Degenhardt F., Bujanda L., Buti M., Albillos A., Invernizzi P. et al. 2020 Genomewide association study of severe covid-19 with respiratory failure. N. Engl. J. Med. https://doi.org/10.1056/nejmoa2020283.
Franceschi C. and Campisi J. 2014 Chronic inflammation (Inflammaging) and its potential contribution to age-associated diseases. J. Gerontol. 69, S4–S9.
Fulop T., Witkowski J. M., Olivieri F. and Larbi A. 2018 The integration of inflammaging in age-related diseases. Semin. Immunol. 40, 17–35.
Gavazzi G. and Krause K. H. 2002 Ageing and infection. Lancet Infect. Dis. 2, 659–666.
Gordon D. E., Jang G. M., Bouhaddou M., Xu J., Obernier K., White K. M., O’Meara M. J. et al. 2020 A SARS-CoV-2 protein interaction map reveals targets for drug repurposing. Nature 583, 459–468.
Harapan H., Itoh N., Yufika A., Winardi W., Keam S., Te H. et al. 2020 Coronavirus disease 2019 (COVID-19): A literature review. J. Infect. Public Heal. 13, 667–673.
Hauser A., Counotte M. J. J., Margossian C. C. C., Konstantinoudis G., Low N. et al. 2020 Estimation of SARS-CoV-2 mortality during the early stages of an epidemic: a modelling study in Hubei, China and northern Italy. MedRxiv. Preprint, https://doi.org/10.1101/2020.03.04.20031104.
Kahan S. M., Wherry E. J., and Zajac A. J. 2015 T cell exhaustion during persistent viral infections. Virology 479–480. 180–193.
Kuleshov M. V., Jones. M. R., Rouillard A. D., Fernandez N. F., Duan Q., Wang Z. et al. 2016 Enrichr: a comprehensive gene set enrichment analysis. Nucleic Acids Res. 44, W90-W97.
López-Otín C., Blasco M. A., Partridge L., Serrano M. and Kroemer G. 2013 The hallmarks of aging. Cell. 153, 1194.
Meiners S., Eickelberg O. and Königshoff M. 2015 Hallmarks of the ageing lung. Eur Respir J, 45, 807–827.
Meyer K. C. 2001 The role of immunity in susceptibility to respiratory infection in the aging lung. Respir. 128, 23–31.
Meyerowitz-Katz G. and Merone L. 2020 A systematic review and meta-analysis of published research data on COVID-19 infection-fatality rates. Medrxiv. Preprint, https://doi.org/10.1101/2020.05.03.20089854.
Panda A., Arjona A., Sapey E., Bai F., Fikrig E., Montgomery R. R. et al. 2009 Human innate immunosenescence: causes and consequences for immunity in old age. Trends Immunol. 30, 325–333.
Peters M. J., Joehanes R., Pilling L. C., Schurmann C., Conneely K. N., Powell J. et al. 2015 The transcriptional landscape of age in human peripheral blood. Nat. Commun. 6, 1–14.
Sanders J. M., Monogue M. L., Jodlowski T. Z. and Cutrell J. B. 2020 Pharmacologic treatments for coronavirus disease 2019 (COVID-19): a review. Jama-J AM Med. Assoc. 323, 1824–1836.
Siordia J. A. 2020 Epidemiology and clinical features of COVID-19: A review of current literature. J. Clin. Virol. 127.
Srinivasan S., Cui H., Gao Z., Liu M., Lu S., Mkandawire W. et al. 2020 Structural genomics of SARS-COV-2 indicates evolutionary conserved functional regions of viral proteins. Viruses 12, 360.
Swain S. L., McKinstry K. K. and Strutt T. M. 2012 Expanding roles for CD4+ T cells in immunity to viruses. Nat. Rev. Immunol. 12, 136–148.
Varga Z., Flammer A. J., Steiger P., Haberecker M., Andermatt R., Zinkernagel A. S. et al. 2020 Endothelial cell infection and endotheliitis in COVID-19. Lancet 395, 1417–1418.
Wu J. T., Leung K., Bushman M., Kishore N., Niehus R., de Salazar P. M. et al. 2020 Estimating clinical severity of COVID-19 from the transmission dynamics in Wuhan, China. Nat. Med. 26, 506–510.
Wu Z. and McGoogan J. M. 2020 Characteristics of and important lessons from the coronavirus disease 2019 (COVID-19) outbreak in China: Summary of a Report of 72314 Cases from the Chinese Center for Disease Control and Prevention. Jama-J AM Med. Assoc. 323, 1239–1242.
Xiong Y., Liu Y., Cao L., Wang D., Guo M., Jiang A. et al. 2020 Transcriptomic characteristics of bronchoalveolar lavage fluid and peripheral blood mononuclear cells in COVID-19 patients. Emerg. Microbes Infect. 9, 761–770.
Yang J., Huang T., Petralia F., Long Q., Zhang B., Argmann C. et al. 2015 Synchronized age-related gene expression changes across multiple tissues in human and the link to complex diseases. Sci. Rep. 5, 1–16.
Zhou Z., Ren L., Zhang L., Zhong J., Xiao Y., Jia Z. et al. 2020 Heightened innate immune responses in the respiratory tract of COVID-19 patients. Cell Host Microbe. 27, 883–890.
Acknowledgments
Central Instrumentation Facility, University of Delhi South Campus provided the computational facility. Special Assistance Programme and Department of Science and Technology, New Delhi, through FIST and DU-DST PURSE programmes to the Department of Genetics, UDSC, Non-NET Fellowship from UGC; and Senior Research Fellowship from Indian Council of Medical Research, New Delhi to UB are gratefully acknowledged. The authors have declared that there are no conflicts of interest in relation to the subjects of this study.
Author information
Authors and Affiliations
Corresponding author
Additional information
Corresponding editor: H. A. Ranganath
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
BHATTACHARYYA, U., THELMA, B.K. Age-related gene expression alterations by SARS-CoV-2 infection contribute to poor prognosis in elderly. J Genet 99, 80 (2020). https://doi.org/10.1007/s12041-020-01233-7
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s12041-020-01233-7