Abstract
Purpose
JARID2, located on chromosome 6p22.3, is a regulator of histone methyltransferase complexes that is expressed in human neurons. So far, 13 individuals sharing clinical features including intellectual disability (ID) were reported with de novo heterozygous deletions in 6p22–p24 encompassing the full length JARID2 gene (OMIM 601594). However, all published individuals to date have a deletion of at least one other adjoining gene, making it difficult to determine if JARID2 is the critical gene responsible for the shared features. We aim to confirm JARID2 as a human disease gene and further elucidate the associated clinical phenotype.
Methods
Chromosome microarray analysis, exome sequencing, and an online matching platform (GeneMatcher) were used to identify individuals with single-nucleotide variants or deletions involving JARID2.
Results
We report 16 individuals in 15 families with a deletion or single-nucleotide variant in JARID2. Several of these variants are likely to result in haploinsufficiency due to nonsense-mediated messenger RNA (mRNA) decay. All individuals have developmental delay and/or ID and share some overlapping clinical characteristics such as facial features with those who have larger deletions involving JARID2.
Conclusion
We report that JARID2 haploinsufficiency leads to a clinically distinct neurodevelopmental syndrome, thus establishing gene–disease validity for the purpose of diagnostic reporting.
Similar content being viewed by others
INTRODUCTION
The JARID2 (jumonji, AT rich interactive domain 2; OMIM 601594) gene is located on chromosome 6p22.3 and encodes a protein that regulates the activity of various histone methyltransferase complexes.1,2,3 JARID2 forms a complex together with polycomb repressive complex 2 (PRC2) that is essential to recruit polycomb group proteins to its target genes. PRC2 can lower gene transcription by catalyzing the di- and trimethylation of lysine 27 on histone H3 (H3K27me2/3). By the regulation of epigenetic changes, the JARID2-PCR2 complex is necessary to control development, differentiation, and survival of embryonic cells.4,5 JARID2 also regulates pluripotency and embryonic stem cell differentiation through Nanog expression and β-catenin.6 In addition, JARID2 has an important function in the Notch-1 pathway, which is essential for development of the central nervous system and other tissues.7 By the methylation of H3-K9 and repression of cyclin D1, JARID2 also regulates cardiomyocytes proliferation and migration of neural progenitor cells.8
JARID2 is crucial in embryogenesis and morphogenesis, and multiple malformations can arise from its dysregulation in mice. In the mouse, Jarid2 is involved in the development of the cardiovascular system, the liver, in hematopoiesis and in neural tube fusion.9 In human embryogenesis, JARID2 is expressed in neurons, especially in the dorsal root ganglion, and in adults it is expressed in the neurons of the cerebral cortex.10
De novo coding single-nucleotide polymorphisms in JARID2 have been found once per study in two autism studies11,12 (p.Arg827Gln and p.Met1181LeufsTer3) and once in a schizophrenia study13 (p.Gly769Ser). However, these single findings did not reach significance in those large studies.
In another study, JARID2 was found to be in linkage disequilibrium with nonsyndromic cleft lip and/or palate. Mouse models showed that Jarid2 is expressed in the merging palatal shelves at the time of fusion, supporting its involvement in palatal development.14 A more recent case–control study found that a deep-intronic JARID2 single-nucleotide variant was protective for nonsyndromic cleft lip and/or palate in a Brazilian cohort.15
Chromosomal deletions in 6p22–p24 involving JARID2 have been identified by karyotype16,17,18,19 and chromosome microarray analysis20,21,22 in 15 individuals, of whom 13 have a complete deletion of JARID2. These individuals have a common phenotype of borderline intellectual functioning to severe intellectual disability (ID) and share characteristic facial features. These features include prominent supraorbital ridges, deep set eyes, infraorbital dark circles, and midface hypoplasia. Apart from JARID2, all of the reported deletions involve other neighboring genes as well (henceforth referred to as JARID2-plus deletions), which has complicated the identification of the critical gene(s). Based on the smallest region of overlap (involving the genes JARID2 and DTNBP1) in four individuals with de novo 6p22.3-24.1 deletions, it has been proposed that JARID2 is a likely candidate gene contributing to the phenotype. This was supported by the finding that JARID2 expression in leukocytes is significantly reduced in these individuals compared with controls.20 Because of the characteristic facial appearance in these individuals, Baroy et al. propose that JARID2 haploinsufficiency may represent a clinically recognizable neurodevelopmental syndrome.20
We describe 16 individuals with developmental delay and/or ID and overlapping clinical features with a deletion or single-nucleotide variant of JARID2. Seven individuals have partial deletions of JARID2 that are predicted to lead to nonsense-mediated messenger RNA (mRNA) decay and one individual has a complete deletion of JARID2. Five individuals have a single-nucleotide variant in JARID2 that leads to a frameshift, stop codon, or splice site alteration, and three individuals have a missense variant. We thus confirm that JARID2 haploinsufficiency leads to a clinically distinct neurodevelopmental syndrome.
MATERIALS AND METHODS
Subjects
Sixteen individuals from 15 unrelated families with a JARID2 deletion or single-nucleotide variant were identified in a diagnostic setting. A collaboration to further analyze and report these cases was established through GeneMatcher, an online platform that facilitates connections between clinicians and researchers who share an interest in the same gene.23 Clinical information was collected by reviewing the medical records. The characteristics of these individuals were compared to evaluate if there was a common phenotype.
Ethics statement
Approval to share clinical and genetic information was received from local institutional review boards (including the IRB of CHU Sainte-Justine and Medical Research Ethics Committee of Amsterdam UMC). Informed consent to publish clinical data was obtained from all families. For individuals where pictures are shown, a signed consent for the publication of photographs was obtained.
Microarray analysis
Comparative genomic hybridization (CGH) array and single-nucleotide polymorphism (SNP) array were performed independently at different centers. CGH array was performed on an Agilent 180K oligoarray in individual 1 and her parents and on an Agilent 105K oligoarray in individual 2 and her parents. CGH array was performed in individual 3 and his parents on an Oxford Gene Technology (OGT) 180K oligonucleotide platform. For individual 4 and his parents, SNP array was performed using an Illumina HumanCytoSNP-12 (v2.1) BeadChip. Illumina CytoSNP-850k SNP array was performed in individual 5 and her father (individual 6), mother, and brother. For individual 7, an oligo-SNP array was performed with Affymetrix CytoScan HD. SNP array with Illumina CytoSNP-12 (v2.1) was performed for individual 8, with parental microarrays performed on an Illumina Infinium Global Screening Array-24 (v2.0) kit.
Exome sequencing
Individual 9 had a commercial Autism/ID Xpanded Panel based on exome capture done at GeneDx lab. This panel uses a trio approach and includes more than 2300 genes associated with autism spectrum disorder and/or ID. Individual 10 had proband-only exome sequencing performed through GeneDx. Individual 11 was enrolled through an IRB-approved research exome sequencing protocol. The process for variant filtering and variant prioritization has been previously described.24,25 Trio-based exome sequencing was completed with clinical confirmation by Sanger sequencing of the JARID2 variant. Individual 12 underwent trio-based exome sequencing as part of a research study (CAUSES Study, approved by University of British Columbia [REB#H15-00092]). Sequencing was performed at Ambry Genetics on an Illumina platform and analysis was performed by the research team at University of British Columbia. Individual 13 had solo exome sequencing performed with an in-house pipeline.26 Parental inheritance was assessed through Sanger sequencing. Individual 14 had trio exome sequencing performed clinically at the Children’s Hospital of Philadelphia. Exons were captured with the Agilent SureSelect XT Clinical Research Exome Version 1 kit (per manufacturer’s protocol) and sequenced on the Illumina HiSeq 2500 platform. Sequencing data were processed using an in-house custom-built bioinformatics pipeline.27,28,29 Individual 15 had a clinical diagnostic exome done with an in-house protocol30 and her parents were assessed only for the variants identified. Individual 16 also had a clinical exome performed with the same protocol as individual 15.30
RESULTS
Clinical characteristics
We identified 4 females and 12 males with a median age of 9.5 years old (range 3.2 to 39 years) with a deletion or single-nucleotide variant in JARID2.
Development and behavior
All individuals have various degrees of developmental delay. Mild to moderate ID was diagnosed in 11/15 (73%) of them. Three individuals had borderline intellectual functioning and one had learning difficulties. Features of autism are noted in more than half of the cohort (9/16 [56%]) and a formal diagnosis of autism spectrum disorder was established in three of these individuals. Behavior abnormalities are present in 7/16 individuals (44%) and include an aggressive demeanor, tendency to obsessive/compulsive and perseverative behavior, attention deficit–hyperactivity disorder (ADHD), and trouble with socialization. Rare manifestations that are only observed in one individual include phonic processing disorder, speech sound disorder, motor dyspraxia, severe stutter, and developmental coordination disorder. One individual also presents two psychotic episodes at the age of 16 years (Table 1, Supplementary table 3).
Neurologic manifestations
Gait disturbance in individuals with JARID2-plus deletions was reported in the past by Baroy et al.20 and Di Benedetto et al.22 but we only identified one individual with a clumsy gait and frequent tripping in our cohort. Hypotonia is found in 5/16 individuals (31%) and only one individual has bradykinesia and bradyphrenia. We identified epilepsy in 3/16 individuals (19%) of the cohort. One individual developed acute epileptic encephalopathy at around age 2 years. Another individual has refractory focal epilepsy and absences. The third individual had epilepsy that resolved at 3 years of age. Nine individuals have been evaluated by brain magnetic resonance image (MRI) or computed tomography (CT) scan. Four individuals have various constitutional anomalies, including benign external hydrocephalus, posterior fossa cyst/mega cisterna magna, periventricular hyperintensities, and arachnoid cyst, but no consistent finding is observed (Table 1, Supplementary table 3).
Dysmorphism
Dysmorphic facial features are observed in 15/16 individuals (94%) (Fig. 1, Supplementary table 3). Dysmorphisms that are observed in more than two individuals are presented in Table 1. The most common features are a high anterior hairline and deep set eyes (6/16 individuals [38%]). Full lips are found in 5/16 (31%) individuals and a broad forehead, infraorbital dark circles, bulbous nasal tip, or depressed nasal bridge in 4/16 individuals (25%). Other less frequently identified dysmorphisms include prominent supraorbital ridges, midface hypoplasia, and a short philtrum (3/6 individuals [19%]). Abnormalities involving hands or feet are found in 5/16 individuals (31%) and include pes planus, clinodactyly of the 4th and 5th toes, persistent fetal pads, single palmar crease, camptodactyly of the 5th digit, syndactyly of the 2nd and 3rd toes, and tapering of the fingers.
Individual 1 (a), individual 2 (b), individual 3 (c), individual 5 (d), individual 6 (who is the father of individual 5) (e), individual 7 (f), individual 11 (g), individual 14 (h), and individual 16 (i). Some individuals share physical features similar to others in the literature with JARID2-plus deletions, including high anterior hairline, broad forehead, deep set eyes, infraorbital dark circles, depressed nasal bridge, bulbous nasal tip and full lips.
Other
Several individuals have had perinatal complications, such as neonatal hyperbilirubinemia (three individuals) and neonatal feeding problems (two individuals). There are five individuals that have a tall stature and four individuals are overweight. One individual has microcephaly, while two have macrocephaly. Only one individual has a cardiac anomaly (tricuspid regurgitation). Musculoskeletal anomalies are observed in five individuals: three individuals have joint hyperlaxity, one has scoliosis, and one has congenital torticollis. Dental anomalies are seen in two individuals: one had hypodontia and the other prominent upper central incisors and irregularly spaced teeth. One individual has a bifid uvula and a submucous cleft palate. Cutaneous findings are inconsistent throughout the cohort. One individual has café au lait macules, one has acanthosis nigricans in the neck and axillae (secondary to obesity) with hirsutism, and another has a patch of prominent capillaries on the upper back. Refractory errors and strabismus are noted in four individuals. There are no individuals with hearing impairment or inner ear anomalies (Table 1, Supplementary table 3).
Genetic variants
Deletions
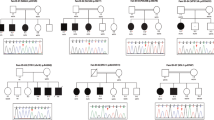
Microarray analysis revealed whole or partial deletions of JARID2 in eight individuals (Figs. 2 and 3, Table 2). All deletions occurred de novo or were inherited from an affected parent, although for two individuals inheritance was not determined. Two de novo deletions were identified that involve only exon 2 of JARID2 (individuals 1 and 3) and two that involve exon 2 and 3 (individuals 2 and 4). Individual 5 was found to have a 140-kb deletion comprising exons 2–5 of JARID2. Her father (individual 6) has a similar deletion, with differences in breakpoints due to inherent measurement uncertainty of the array platform. The error margins of their breakpoints lie fully within the intronic region. The mother of individual 5 has a normal female microarray profile and the healthy brother of individual 5 has a normal targeted array for the familial deletion. Individual 7 has a deletion that includes exons 1 and 2 of JARID2. Individual 8 has a de novo deletion encompassing all of JARID2 and the distal end of DTNBP1 (involving the last three exons).
Deletions reported in this study (a) and JARID2-plus deletions previously reported by Di Benedetto et al.22 (b), Baroy et al.20 (c), Celestino-Soper et al.21 (d) and deletions from Swaay et al.19, Davies et al.,16 Davies et al.,17 and Zirn et al.18 as labeled by Celestino-Soper et al.21 (e). Blue shade covers the JARID2 gene region. For some of the reported individuals exact breakpoints were not given: the minimal size of the deleted region is presented in this figure. Data were uploaded into UCSC Genome Browser (genome.ucsc.edu).
The intragenic JARID2 deletions are likely to result in a frameshift that will lead to a premature stop codon. The predicted effect would be a loss of normal protein function through nonsense-mediated mRNA decay. Complete deletion of JARID2, as identified in one individual, is predicted to be pathogenic.
Single-nucleotide variants
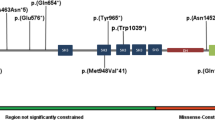
We identified single-nucleotide variants of JARID2 in eight individuals (Fig. 3 and Table 2). Two de novo frameshift variants were identified (individuals 9 and 11). Two individuals have a nonsense variant, of which one is de novo (individual 12). For the other one inheritance could not be determined because of adoption (individual 10). One individual (individual 13) has a de novo variant c.2731+1G>C that is predicted to affect splicing since it affects a canonical splice site nucleotide. However, functional testing was not performed. These five variants are predicted to be pathogenic and lead to protein loss of function due to a splicing aberration or nonsense-mediated mRNA decay.
There are three individuals with a de novo missense variant (individuals 14, 15, and 16). The missense variants affect highly conserved residues as shown in Fig. 3. Pathogenicity predictions for missense and splice site variants are shown in Supplementary table 1. Multiple pathogenicity prediction tools classified missense variants as pathogenic; all were considered pathogenic by DANN, FATHMM-MKL, MutationTaster, and SIFT although other tools predicted they were benign. They all had CADD scores above 20 (26.5, 31, and 24.6, respectively), which means they are classified among the top 1% of variants in the genome with respect to pathogenicity probability.
Individuals with a different phenotype or other explanatory variants
We identified two other individuals with deletion or single-nucleotide variation in JARID2 but they presented with a different phenotype or had other variations that could explain their phenotype. One individual with rhabdomyolysis had a de novo missense variant (c.3362A>G, p.[Asp1121Gly]) in JARID2. Another individual with a de novo missense variant in JARID2 (c.2480G>A, p.[Arg827Gln]) was reported with a phenotype similar to our patients. The individual had ID, global developmental delay, autistic features, hypotonia, pes planus, and delayed myelination on MRI. He also had short stature, dysplastic semicircular canals, cardiac anomalies, feeding and breathing difficulties at birth, and some dysmorphisms that were not overlapping those of our patients (telecanthus, epicanthal folds, narrow palpebral fissures, broad nose, and long philtrum). Trio exome sequencing showed another de novo variant in TLK2 (c.887T>C, p.[Leu296Pro]). This variant was further reclassified as likely pathogenic by the diagnostic laboratory and is currently the main candidate to explain the clinical phenotype. We are uncertain if the JARID2 variant contributes to or exacerbates the phenotype, so we did not include this individual in our previous analyses. Bioinformatic predictions of these variants, as well as variants that were previously reported in the literature, are presented in Supplementary table 2.
DISCUSSION
We describe 16 individuals from 15 families with a deletion or single-nucleotide variant of JARID2. All individuals described in this paper have developmental delay and the majority have ID. The four individuals without ID however have borderline intellectual functioning and/or learning difficulties. Other common characteristics include hypotonia, autistic features, and behavior abnormalities, especially aggressive behavior. In some patients, we report similar physical features to previously reported cases in the literature with JARID2-plus deletions, including high anterior hairline, broad forehead, deep set eyes, infraorbital dark circles, depressed nasal bridge, bulbous nasal tip, and full lips. Patients with deletions tend to have more overlapping facial features than individuals with missense variants. This may be because missense variants cause a more moderate loss-of-function effect on JARID2. Our cohort is not large enough to determine if this trend is significant.
The identified JARID2 deletions are predicted to lead to a loss of normal protein function, as well as the frameshift, nonsense, and splice site variants that were detected. Hence, these cases confirm the hypothesis by Barøy et al. that it is JARID2 haploinsufficiency that leads to a clinically distinct neurodevelopmental syndrome.
It is noteworthy that in one case (individual 5) the JARID2 deletion was inherited from an affected parent (individual 6). As some individuals only have a mild developmental delay or borderline intellectual functioning, we expect further patients to be identified with a pathogenic JARID2 variant inherited from a mildly affected parent. There are no segmental duplications within JARID2 that could explain the potentially recurrent breakpoint within intron 1, but there are Alu sequences that could potentially mediate Alu/Alu recombination.
Thus far, there has only been one other report of de novo intragenic JARID2 deletions. That study described five individuals with ID and de novo intragenic JARID2 deletions (as well as two duplications), all of them involving only exon 6 (exon 5 in NM_004973.4, 177 nucleotides).31 Further in silico investigation showed that heterozygous loss or gain of JARID2 exon 6 does not predict a frameshift and is likely to be tolerated. Additionally, they found a high frequency (>14%) of JARID2 exon 6 copy-number variants (CNVs) in control populations.32 The authors therefore concluded that these CNVs are unlikely to be causative for ID, although they might have a contributory effect. The JARID2 deletions in our patients, however, were predicted to lead to a frameshift and no comparable losses were found in control populations reported in the Database of Genomic Variants (DGV, http://dgv.tcag.ca, accessed 26 May 2020).
The DGV contains only one individual with a JARID2-plus deletion (deletion of exons 7–18 of JARID2 and a partial deletion of the adjacent gene DTNBP1).33 This partial deletion of JARID2 and DTNBP1 is similar to the deletion identified in one of the patients reported by Baroy et al. (Fig. 2, B4)20 who had an IQ of 74. Possibly, healthy population databases might contain data on people with borderline intellectual functioning. One other exonic deletion is reported in the DGV that encompasses only exon 1 of JARID2.34 There are no deletions that involve all of JARID2 in the DGV. Finally, a small deletion (133 kb) involving the first three exons of JARID2 was previously reported in an individual with isolated talipes equinovarus and his unaffected father who were reported to be cognitively normal and without a history of developmental delay (Gurnett, personal communication).35
Interestingly, there is one previous report of an individual with a de novo probably pathogenic missense variant in JARID2 (c.2255C>T, p.[Pro752Leu]) from a cohort of 92 patients with syndromic ID.36 Although no pathogenic JARID2 single-nucleotide variants were described previously, pathogenic variants in other members of the JmjC domain–containing family of proteins have been associated with human diseases, including neurodevelopmental disorders.37,38,39,40 Because JARID2 bears most resemblance to JARID1 proteins, pathogenic variants in KDM5C (JARID1C, OMIM 314690), associated with X-linked ID (OMIM 300534), and pathogenic variants in KDM5B (JARID1B, OMIM 605393), causing a form of autosomal recessive ID (OMIM 618109), are of most interest. In addition to the JmjC domain, these JARID1 proteins contain a Jumonji N (JmjN) domain, AT rich interaction domain (ARID), and a zinc finger (ZF) as well.6
Furthermore, expected and observed counts of single-nucleotide changes in gnomAD show that JARID2 is extremely intolerant to loss-of-function variants (probability of loss of function intolerance [pLI] score 1; observed/expected [o/e] ratio 0.09, 90% confidence interval [CI]: 0.05–0.19). Also, fewer missense variants are observed than expected (o/e ratio 0.73 [90% CI: 0.68–0.78] with a Z-score of 2.69) (https://gnomad.broadinstitute.org/, accessed 20 May 2020). Regarding further bioinformatic analysis of JARID2 as a dominant disease gene, the %HI score (from DECIPHER) is 12.14%. High %HI ranks (e.g., 0–10%) indicate a gene is more likely to exhibit haploinsufficiency. The JARID2 P(AD) score is 0.996 (from DOMINO, wwwfbm.unil.ch/domino, accessed 20 May 2020). A P(AD) score of ≥0.95 is highly associated with autosomal dominant inheritance through haploinsufficiency, gain-of-function, or dominant-negative effects.41
Conclusion
We propose that JARID2 should be considered as a critical gene in the 6p22–p24 region with haploinsufficiency resulting in developmental delay and/or borderline intellectual functioning to severe intellectual disability. In addition to JARID2 deletions, loss-of-function single-nucleotide variants in this gene result in a similar neurodevelopmental syndrome. Currently, there are only three tests available in the Genetic Testing Registry that offer JARID2 sequencing (https://www.ncbi.nlm.nih.gov/gtr/all/tests/?term=jarid2, accessed 14 May 2020). Our data provide further evidence for establishing gene–disease validity for the purpose of diagnostic reporting and we suggest adding JARID2 to ID gene panels.
In summary, we propose that haploinsufficiency of JARID2 be considered as a new, clinically distinct neurodevelopmental syndrome.
Change history
22 October 2020
The original online PDF version of the Article contained figures in monochrome. They now appear in colour in the PDF and HTML versions of the Article.
References
Li G, Margueron R, Ku M, Chambon P, Bernstein BE, Reinberg D. Jarid2 and PRC2, partners in regulating gene expression. Genes Dev. 2010;24:368–380.
Mysliwiec MR, Carlson CD, Tietjen J, Hung H, Ansari AZ, Lee Y. Jarid2 (Jumonji, AT rich interactive domain 2) regulates NOTCH1 expression via histone modification in the developing heart. J Biol Chem. 2012;287:1235–1241.
Pasini D, Cloos PA, Walfridsson J, et al. JARID2 regulates binding of the Polycomb repressive complex 2 to target genes in ES cells. Nature. 2010;464:306–310.
Shen X, Kim W, Fujiwara Y, et al. Jumonji modulates polycomb activity and self-renewal versus differentiation of stem cells. Cell. 2009;139:1303–1314.
Peng JC, Valouev A, Swigut T, et al. Jarid2/Jumonji coordinates control of PRC2 enzymatic activity and target gene occupancy in pluripotent cells. Cell. 2009;139:1290–1302.
Landeira D, Bagci H, Malinowski AR, et al. Jarid2 coordinates nanog expression and PCP/Wnt signaling required for efficient ESC differentiation and early embryo development. Cell Rep. 2015;12:573–586.
Yoon K, Gaiano N. Notch signaling in the mammalian central nervous system: insights from mouse mutants. Nat Neurosci. 2005;8:709–715.
Shirato H, Ogawa S, Nakajima K, et al. A jumonji (Jarid2) protein complex represses cyclin D1 expression by methylation of histone H3-K9. J Biol Chem. 2009;284:733–739.
Jung J, Mysliwiec MR, Lee Y. Roles of JUMONJI in mouse embryonic development. Dev Dyn. 2005;232:21–32.
Berge-Lefranc JL, Jay P, Massacrier A, et al. Characterization of the human jumonji gene. Hum Mol Genet. 1996;5:1637–1641.
Yuen RK, Merico D, Cao H, et al. Genome-wide characteristics of de novo mutations in autism. NPJ Genom Med. 2016;1:160271–1602710.
De Rubeis S, He X, Goldberg AP, et al. Synaptic, transcriptional and chromatin genes disrupted in autism. Nature. 2014;515:209–215.
Fromer M, Pocklington AJ, Kavanagh DH, et al. De novo mutations in schizophrenia implicate synaptic networks. Nature. 2014;506:179–184.
Scapoli L, Martinelli M, Pezzetti F, et al. Expression and association data strongly support JARID2 involvement in nonsyndromic cleft lip with or without cleft palate. Hum Mutat. 2010;31:794–800.
Messetti AC, Machado RA, de Oliveira CE, et al. Brazilian multicenter study of association between polymorphisms in CRISPLD2 and JARID2 and nonsyndromic oral clefts. J Oral Pathol Med. 2017;46:232–239.
Davies AF, Olavesen MG, Stephens RJ, et al. A detailed investigation of two cases exhibiting characteristics of the 6p deletion syndrome. Hum Genet. 1996;98:454–459.
Davies AF, Mirza G, Sekhon G, et al. Delineation of two distinct 6p deletion syndromes. Hum Genet. 1999;104:64–72.
Zirn B, Hempel M, Hahn A, et al. Polyneuropathy, scoliosis, tall stature, and oligodontia represent novel features of the interstitial 6p deletion phenotype. Am J Med Genet A. 2008;146:2960–2965.
van Swaay E, Beverstock GC, van de Kamp JJ. A patient with an interstitial deletion of the short arm of chromosome 6. Clin Genet 1988;33:95–101.
Baroy T, Misceo D, Stromme P, et al. Haploinsufficiency of two histone modifier genes on 6p22.3, ATXN1 and JARID2, is associated with intellectual disability. Orphanet J Rare Dis. 2013;8:3.
Celestino-Soper PB, Skinner C, Schroer R, et al. Deletions in chromosome 6p22.3-p24.3, including ATXN1, are associated with developmental delay and autism spectrum disorders. Mol Cytogenet. 2012;5:17.
Di Benedetto D, Di Vita G, Romano C, et al. 6p22.3 deletion: report of a patient with autism, severe intellectual disability and electroencephalographic anomalies. Mol Cytogenet. 2013;6:4.
Sobreira N, Schiettecatte F, Valle D, Hamosh A. GeneMatcher: a matching tool for connecting investigators with an interest in the same gene. Hum Mutat. 2015;36:928–930.
Petrovski S, Aggarwal V, Giordano JL, et al. Whole-exome sequencing in the evaluation of fetal structural anomalies: a prospective cohort study. Lancet. 2019;393:758–767.
Zhu X, Petrovski S, Xie P, et al. Whole-exome sequencing in undiagnosed genetic diseases: interpreting 119 trios. Genet Med. 2015;17:774–781.
Nambot S, Thevenon J, Kuentz P, et al. Clinical whole-exome sequencing for the diagnosis of rare disorders with congenital anomalies and/or intellectual disability: substantial interest of prospective annual reanalysis. Genet Med. 2018;20:645–654.
Baker SW, Murrell JR, Nesbitt AI, et al. Automated clinical exome reanalysis reveals novel diagnoses. J Mol Diagn. 2019;21:38–48.
Gibson KM, Nesbitt A, Cao K, et al. Novel findings with reassessment of exome data: implications for validation testing and interpretation of genomic data. Genet Med. 2018;20:329–336.
Wu C, Devkota B, Evans P, et al. Rapid and accurate interpretation of clinical exomes using Phenoxome: a computational phenotype-driven approach. Eur J Hum Genet. 2019;27:612–620.
Lelieveld SH, Reijnders MR, Pfundt R, et al. Meta-analysis of 2,104 trios provides support for 10 new genes for intellectual disability. Nat Neurosci. 2016;19:1194–1196.
Tucker T, Zahir FR, Griffith M, et al. Single exon-resolution targeted chromosomal microarray analysis of known and candidate intellectual disability genes. Eur J Hum Genet. 2014;22:792–800.
Zahir FR, Tucker T, Mayo S, et al. Intragenic CNVs for epigenetic regulatory genes in intellectual disability: survey identifies pathogenic and benign single exon changes. Am J Med Genet A. 2016;170:2916–2926.
Shaikh TH, Conlin LK, Geiger EA, et al. High-resolution mapping and analysis of copy number variations in the human genome: a data resource for clinical and research applications. Genome Res. 2009;19:1682–1690.
Wong KK, deLeeuw RJ, Dosanjh NS, et al. A comprehensive analysis of common copy-number variations in the human genome. Am J Hum Genet. 2007;80:91–104.
Alvarado DM, Buchan JG, Frick SL, Herzenberg JE, Dobbs MB, Gurnett CA. Copy number analysis of 413 isolated talipes equinovarus patients suggests role for transcriptional regulators of early limb development. Eur J Hum Genet. 2013;21:373–380.
Martinez F, Caro-Llopis A, Rosello M, et al. High diagnostic yield of syndromic intellectual disability by targeted next-generation sequencing. J Med Genet. 2017;54:87–92.
Abidi F, Miano M, Murray J, Schwartz C. A novel mutation in the PHF8 gene is associated with X-linked mental retardation with cleft lip/cleft palate. Clin Genet. 2007;72:19–22.
Adam MP, Banka S, Bjornsson HT, et al. Kabuki syndrome: international consensus diagnostic criteria. J Med Genet. 2019;56:89–95.
Stolerman ES, Francisco E, Stallworth JL, et al. Genetic variants in the KDM6B gene are associated with neurodevelopmental delays and dysmorphic features. Am J Med Genet A. 2019;179:1276–1286.
Najmabadi H, Hu H, Garshasbi M, et al. Deep sequencing reveals 50 novel genes for recessive cognitive disorders. Nature. 2011;478:57–63.
Quinodoz M, Royer-Bertrand B, Cisarova K, Di Gioia SA, Superti-Furga A, Rivolta C. DOMINO: using machine learning to predict genes associated with dominant disorders. Am J Hum Genet. 2017;101:623–629.
Acknowledgements
We thank the families described in this study. Individual 12 was enrolled in the CAUSES Study; investigators include Shelin Adam, Christele Du Souich, Alison Elliott, Anna Lehman, Jill Mwenifumbo, Tanya Nelson, Clara Van Karnebeek, and Jan Friedman; it is funded by Mining for Miracles, British Columbia Children’s Hospital Foundation (grant number F15-01355) and Genome British Columbia (grant number F16-02276). P.M.C. is supported by awards from the Canadian Institutes of Health Research and the Fonds de la Recherche du Quebec–Santé.
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Disclosure
The authors declare no conflicts of interest.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
About this article
Cite this article
Verberne, E.A., Goh, S., England, J. et al. JARID2 haploinsufficiency is associated with a clinically distinct neurodevelopmental syndrome. Genet Med 23, 374–383 (2021). https://doi.org/10.1038/s41436-020-00992-z
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41436-020-00992-z
Keywords
This article is cited by
-
RENGE infers gene regulatory networks using time-series single-cell RNA-seq data with CRISPR perturbations
Communications Biology (2023)