Abstract
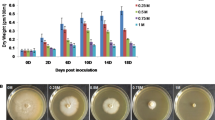
Isatis indigotica Fort. is extremely widely planted and used for the source of the traditional Chinese medicine Radix Isatidis (Ban-Lan-Gen), which had pharmacological activities such as anti-endotoxic, antibacterial, antinociceptive, anti-inflammatory and antipyretic. In the cultivation, salt stress could affect the yields of Radix Isatidis, and activate the secondary metabolism, eg. the contents of epigoitrin in root and indigo in leaf increased after a period of salt stress, which was possibly regulated by related genes. But in early salt stress, the molecular response of I. indigotica and expression of related genes are also worthy of our attention, which it helps us to more fully understand the molecular response mechanism of I. indigotica, so that we can make better use of “salt stress” to stimulate its secondary metabolism. To determine the molecular changes of I. indigotica in salt stress, the RNA-seq was carried out for the expression profile with NGS QC Toolkit (v2.3.2) software, the Illumina HiSeqTM 2000 in Biomarker Technologies Co., Ltd and ESTScan program. The de novo assembly resulted in 33109 unigenes from more than 18.71 Gb data. Of these, 28868 unigenes were annotated using KOG, KEGG, Nr, Nt, Swiss-Prot and TrEMBL databases. Then, 11829 simple sequence repeats were found in unigenes and 7725 primers were designed. After detecting the expression value, the edgeR package found 135 DEG, of those 48 were up-regulated and 87 were down-regulated. The expression pattern of genes that was validated by qPCR indicated that mitochondrial transcription termination factor, non-LTR retroelement reverse transcriptase and CYP79F1 which involved in the first step in biosynthesis of core short-chain aliphatic glucosinolates might play a vital role in coping with salt stress. Although one assembly of I. indigotica unigenes were obtained before, this study would provide more molecular data for further analysis, and facilitate studies on the functions of genes involved in the salt related signal pathways.



Similar content being viewed by others
REFERENCES
Chinese Pharmacopoeia Commission. Pharmacopoeia of the People’s Republic of China, Beijing: China medical science press, 2015.
Kong, W.J., Zhao, Y.L., Shan, L.M., Xiao, X.H., and Guo, W.Y., Investigation of the effect of four organic acids in Radix isatidis on E. coli growth by microcalorimetry, Chin. J. Chem., 2008, vol. 26, p. 113.
Ho, Y.L. and Chang, Y.S., Studies on the antinociceptive, anti-inflammatory and antipyretic effects of Isatis indigotica root, Phytomedicine, 2002, vol. 9, p. 419.
Zhou, J., van Staden, J., Guo, L.P., and Huang, L.Q., Smoke-water improves shoot growth and indigo accumulation in shoots of Isatis indigotica seedlings, S. Afr. J. Bot., 2011, vol. 77, p. 787.
Han, G.H., Gim, G.H., Kim, W., Seo, S.I., and Kim, S.W., Enhanced indirubin production in recombinant Escherichia coli harboring a flavin-containing monooxygenase gene by cysteine supplementation, J. Biotechnol., 2012, vol. 164, p. 179.
Deng, X.Y., Gao, G.H., Zheng, S.N., and Li, F.M., Qualitative and quantitative analysis of flavonoids in the leaves of Isatis indigatica Fort. by ultra-performance liquid chromatography with PDA and electrospray ionization tandem mass spectrometry detection, J. Pharm. Biomed. Anal., 2008, vol. 48, p. 562.
Xu, T.F., Zhang, L., Sun, X.F., Zhang, H.M., and Tang, K.X., Production and analysis of organic acids in hairy-root cultures of Isatis indigotica Fort. (indigo woad), Biotechnol. Appl. Biochem., 2004, vol. 39, p. 123.
Wang, X., Xie, Y., Hu, X., Li, Y., Hu, P., Wang, Y., Liang, Q., and Luo, G., Qualitative and quantitative analysis of glucosinolates and nucleosides in Radix isatidis by HPLC and liquid chromatography tandem mass spectrometry, Acta Pharma. Sinica B, 2013, vol. 3, p. 337.
Li, T., Qu, X.Y., Zhang, Q.A., and Wang, Z.Z., Ultrasound-assisted extraction and profile characteristics of seed oil from Isatis indigotica Fort., Ind. Crops Prod., 2012, vol. 35, p. 98.
Wang, Y., Zhou, R.Y., Ma, L.M., Bai, Y., Guan, J.L., and Tang, X.Q., Seed germination and seedling growth response of Isatis indigotica in five populations from saline environments, Acta Pratacult. Sin., 2018, vol. 27, p. 145.
Tang, X.Q., Lv, T.T., Zhao, X.L., Jin, M.H., Xiao, Y.H., and Wang, K.C., Effect of spraying ALA on growth and main active component content of Isatis indigotica seedling during NaCl stress process, J. Plant Resour. Environ., 2014, vol. 23, p. 55.
Kawasaki, S., Borchert, C., Deyholos, M., Wang, H., Brazille, S., Kawai, K., Galbraith, D., and Bohnert, H., Gene expression profiles during the initial phase of salt stress in rice, Am. Soc. Plant Physiol., 2001, vol. 13, p. 889.
Tang, X.Q., Xiao, Y.H., Lv, T.T., Wang, F.Q., Zhu, Q.H., Zheng, T.Q., and Yang, J., High-throughput sequencing and de novo assembly of the Isatis indigotica transcriptome, PLoS One, 2014, vol. 9: e102963.
Patel, R.K. and Jain, M., NGS QC Toolkit: a toolkit for quality control of next generation sequencing data, PLoS One, 2012, vol. 7: e30619. https://doi.org/10.1371/journal.pone.0030619
Haas, B.J., Papanicolaou, A., Yassour, M., Grabherr, M., Blood, P.D., Bowden, J., Couger, M.B., Eccles, D., Li, B., Lieber, M., MacManes, M.D., Ott, M., Orvis, J., Pochet, N., Strozzi, F., et al., De novo transcript sequence reconstruction from RNA-seq using the Trinity platform for reference generation and analysis, Nat. Protoc., 2013, vol. 8, p. 1494.
Iseli, C., Jongeneel, C.V., and Bucher, P., ESTScan: a program for detecting, evaluating, and reconstructing potential coding regions in EST sequences, Proc. Int. Conf. Intell. Syst. Mol. Biol., 1999, p. 138.
Ye, J., Fang, L., Zheng, H.K., Zhang, Y., Chen, J., Zhang, Z.J., Wang, J., Li, S.T., Li, R.Q., Bolund, L., and Wang, J., WEGO: a web tool for plotting GO annotations, Nucleic Acids Res., 2006, vol. 34, p. 293.
Robinson, M.D., McCarthy, D.J., and Smyth, G.K., Edge R: a Bioconductor package for differential expression analysis of digital gene expression data, Bi-oinformatics, 2010, vol. 26, p. 139.
Xu, Z.S., Tan, H.W., Wang, F., Hou, X.L., and Xiong, A.S., CarrotDB: a genomic and transcriptomic database for carrot, Journal Biological Databases and Curation, 2014. https://doi.org/10.1093/database/bau096
Lawson, M.J. and Zhang, L.Q., Distinct patterns of SSR distribution in the Arabidopsis thaliana and rice genomes, Genome Biol., 2006, vol. 7: R14.
Singh, H., Deshmukh, R.K., Singh, A., Singh, A.K., Gaikwad, K., Sharma, T.R., Mohapatra, T., and Singh, N.K., Highly variable SSR markers suitable for rice genotyping using agarose gels, Mol. Breed., 2010, vol. 25, p. 359.
Trapnell, C., Williams, B.A., Pertea, G., Mortazavi, A., Kwan, G., van Baren, M.J., Salzberg, S.L., Wold, B.J., and Pachter, L., Transcript assembly and quantification by RNA-Seq reveals unannotated transcripts and isoform switching during cell differentiation, Nat. Biotec-hnol., 2010, vol. 28, p. 511. https://doi.org/10.1038/nbt.1621
Lee, S.H., Lee, J.H., Paek, K.H., Kwon, S.Y., Cho, H.S., Kim, S.J., and Park, J.M., A novel WD40 protein, BnSWD1, is involved in salt stress in Brassica napus,Plant Biotechnol. Rep., 2010, vol. 2, p. 165.
Munns, R. and Tester, M., Mechanisms of salinity tolerance, Annu. Rev. Plant Biol., 2008, vol. 59, p. 651.
Deinlein, U., Stephan, A.B., Horie, T., Luo, W., Xu, G., and Schroeder, J.I., Plant salt-tolerance mechanisms, Trends Plant Sci., 2014, vol. 19, p. 371.
Chen, Y., Lei, S., Zhou, Z., Zeng, F., Yi, B., Wen, J., Shen, J., Ma, C., Tu, J., and Fu, T., Analysis of gene expression profile in pollen development of recessive genic male sterile Brassica napus L. line S45A, Plant Cell Rep., 2009, vol. 28, p. 1363.
Grandbastien, M.A., Activation of plant retrotransposons under stress conditions, Trends Plant Sci., 1998, vol. 3, p. 181.
Robles, P., Micol, J.L., and Quesada, V., Arabidopsis MDA1, a nuclear-encoded protein, functions in chloroplast development and abiotic stress responses, PLoS One, 2012, vol. 7: e42924.
Pang, Q.Y., Guo, J., Chen, S.X., Chen, Y.Z., Zhang, L., Fei, M.H., Jin, S.J., Li, M.S., Wang, Y., and Yan, X.F., Effect of salt treatment on the glucosinolate-myrosinase system in Thellungiella salsuginea,Plant Soil, 2012, vol. 355, p. 363.
Dhaubhadel, S. and Krishna, P., Identification of differentially expressed genes in brassinosteroid-treated Brassica napus seedlings, Plant Growth Regul., 2008, vol. 27, p. 297.
ACKNOWLEDGMENTS
We also thank Nanjing Genepioneer Biotechnologies Co, Ltd., China for help on bioinformatics analysis.
Funding
This study was supported by the National Natural Science Foundation of China (project no. 31171468), National Survey of Traditional Chinese Medicine Resources for the 2018 Chinese Medicine Public Health Service Subsidy (Ministry of Finance and Ministry of Human Resources and Social Security [Caishe 2018] No. 43). The subsidy fund for the improvement of medical services and guarantee capabilities in 2019 (inheritance and development of traditional Chinese medicine) “National Survey of Traditional Chinese Medicine Resources” (Caishe [2019] No. 39). Funds for basic scientific research in central universities (project no. KYZZ201913). Zhenjiang Jinshan Talents in Jiangsu Province of China- Modern Agricultural Leaders (Innovation) Project (2018).
Author information
Authors and Affiliations
Contributions
X.-Q. T., H.-W. T. and J. Y. conceived and designed the experimental plan. S. S. and F. W. participated in sample collection, physiological experiment and RNA preparation. H. T. performed the bioinformatics analysis. X.-Q. T. and H.-W. T. drafted the manuscript. All authors contributed to the revision of this manuscript, and approved the final manuscript.
Corresponding author
Ethics declarations
The authors declare that they have no conflict of interest. This article does not contain any studies involving animals or human participants as objects of research.
Rights and permissions
About this article
Cite this article
Tang, XQ., Tan, HW., Shi, SL. et al. Characterization of Transcriptome Expression: The Response of Isatis indigotica to Salt Stress. Russ J Plant Physiol 67, 1135–1143 (2020). https://doi.org/10.1134/S1021443720060163
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1134/S1021443720060163