Abstract
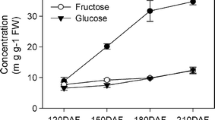
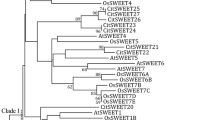
The plant SWEET (Sugars Will Eventually be Exported Transporters) proteins, as newly discovered sugar transporters, play various important roles during plant depending on the transmembrane gradient of sugar concentration. SWEET17 protein is one of the SWEET family members transporting fructose in particular. In this study, we identified eight SWEET17s from six loquats (Eriobotrya japonica (Thunb.) Lindl.) cultivars and named them as Ej_NHB_SWEET17, Ej_BL_SWEET17a, Ej_ZH_SWEET17, Ej_MM-_SWEET17a, Ej_MM_SWEET17b, Ej_BL_SWEET17b, Ej_GY_SWEET17 and Ep_LY_SWEET17. All of the eight loquat SWEET17s have five to six transmembrane structures. Phylogenetic analysis indicated that Ej_NHB_SWEET17 had the farthest genetic relationship with other seven loquat SWEET17s, indicating that ‘Ninghaibai’ cultivar was far away from the other five loquat varieties possibly. The similarities of nucleotide and amino acid sequences between Ej_NHB_SWEET17 and the other loquat SWEET17s were only about 61 and 46%, respectively, while that among other seven SWEET17s were greater than 90%. The expression levels of loquat SWEET17s in leaves were positively correlated with the content of fructose, sorbitol and total sugar in various fruit from the six cultivars, in which the sugar contents in fruits of different cultivars varied significantly, that was probably related to expression levels of loquat SWEET17. Our results indicated that the expression level of loquat SWEET17s was probably related to the regulation of sugar contents of mature fruit, experimental confirmation needs to be performed. Together, these results indicated that EjSWEET17s possibly played important roles in the sugar accumulation of fruit, which could serve as the basis for further functional identification of such genes.





Similar content being viewed by others
REFERENCES
Ma, H., Study on Selective Transformation of Carbohydrates, Guangzhou: South China Univ. Technol., 2015.
Meyer, R.C., Steinfath, M., Lisec. J., Becher, M., Witucka-Wall, H., Törjék, O., Fiehn, O., Eckardt, A., Willmitzer, L., Selbig, J., and Altmann, T., The metabolic signature related to high plant growth rate in Arabi-dopsis thaliana,Proc. Natl. Acad. Sci. USA, 2007, vol. 104, p. 4759.
Zhang, W. and Guo, D.P., Sugar, abscisic acid and ethylene in relation to plant growth and development, J. Plant Physiol., 2005, vol. 41, p. 376.
Smeekens, S., Ma, J., Hanson, J., and Rolland, F., Sugar signals and molecular networks controlling plant growth, Curr. Opin. Plant Biol., 2010, vol. 13, p. 273.
Zhang, Y., Zhang, D.B., and Liu, M., The molecular mechanism of long-distance sugar transport in plants, Chin. Bull. Bot., 2015, vol. 50, p. 107.
Rennie, E.A. and Turgeon, R.A., Comprehensive picture of phloem loading strategies, Proc. Natl. Acad. Sci. USA, 2009, vol. 106, p. 14162.
Chen, L.Q., Hou, B.H., Lalonde, S., Takanaga, H., Hartung, M.L., Qu, X.Q., Guo, W.J., Kim, J.G., Underwood, W., Chaudhuri, B., Chermak, D., Antony, G., White, F., Somerville, S.C., Mudgett, B., et al., Sugar transporters for intercellular exchange and nutrition of pathogens, Nature, 2010, vol. 468, p. 527.
Wormit, A., Trentmann, O., Feifer, I., Lohr, C., Tjaden, J., Meyer, S., and Neuhaus, H.E., Molecular identification and physiological characterization of a novel monosaccharide transporter from Arabidopsis involved in vacuolar sugar transport, Plant Cell, 2006, vol. 18, p. 3476.
Sauer, N., Molecular physiology of higher plant sucrose transporters, FEBS Lett., 2007, vol. 581, p. 2309.
Aluri, S. and Büttner, M., Identification and functional expression of the Arabidopsis thaliana vacuolar glucose transporter 1 and its role in seed germination and flowering, Proc. Natl. Acad. Sci. USA, 2007, vol. 104, p. 2537.
Schneider, S., Hulpke, S., Schulz, A., Yaron, I., Höll, J., Imlau, A., Schmitt, B., Batz, S., Wolf, S., Hedrich, R., and Sauer, N., Vacuoles release sucrose via tonoplast-localised SUC4-type transporters, Plant Biol., 2012, vol. 14, p. 32.
Lalonde, S., Wipf, D., and Frommer, W.B., Transport mechanisms for organic forms of carbon and nitrogen between source and sink, Annu. Rev. Plant Biol., 2004, vol. 55, p. 341.
Chong, J., Piron, M.C., Meyer, S., Merdinoglu, D., Bertsch, C., and Mestre, P., The SWEET family of sugar transporters in grapevine: VvSWEET4 is involved in the interaction with Botrytis cinerea,J. Exp. Bot., 2014, vol. 65, p. 6589.
Yuan, M. and Wang, S.P., Rice Mt N3/Saliva/SWEET family genes and their homologs in cellular organisms, Mol. Plant, 2013, vol. 6, p. 665.
Gao, Y., Wang, Z.Y., Kumar, V., Yuan, P., Zhu, X.F., Li, TY., Jia, B., and Xuan, Y.H., Genome-wide identification of the SWEET gene family in wheat, Gene, 2018, vol. 642, p. 284.
Xuan, Y.H., Hu, Y.B., Chen, L.Q., Sosso, D., Ducat, D.C., Hou, B.H., and Frommer, W.B., Functional role of oligomerization for bacterial and plant SWEET sugar transporter family, Proc. Natl. Acad. Sci. USA, 2013, vol. 110, p. 3685.
Chardon, F., Bedu, M., Calenge, F., Klemensp, A.W., Spinner, L., Clement, G., Chietera, G., Léran, S., Ferrand, M., Lacombe, B., Loudet, O., Dinant, S., Bellini, C., Neuhaus, H.E., Daniel-Vedele, F., et al., Leaf fructose content is controlled by the vacuolar transporter SWEET17 in Arabidopsis,Curr. Biol., 2013, vol. 23, p. 697.
Guo, W.J., Nagy, R., Chen, H.Y., and Pfrunder, S., Yu, YC., Santelia, D., Frommer, W.B., and Martinoia, E., SWEET17, a facilitative transporter, mediates fructose transport across the tonoplast of Arabidopsis roots and leaves, Plant Physiol., 2014, vol. 164, p. 777.
Chen, L.Q., Qu, X.Q., Hou, B.H., Sosso, D., and Osorio, S., Sucrose efflux mediated by SWEET proteins as a key step for phloem transport, Science, 2012, vol. 335, p. 207.
Wei, X.Y., Identification of the Sugar Transporter Gene Families and Study on the Functions of Two Hexose Sugar Transporters in Malus domestica, Northwest A&F University, 2015.
Zhou, A., Ma, H., Feng, S., Gong, S., and Wang, J., DsSWEET17, a tonoplast-localized sugar transporter from Dianthus spiculifolius, affects sugar metabolism and confers multiple stress tolerance in Arabidopsis,Int. J. Mol. Sci., 2018, vol. 19, p. 1564.
Li, J., Qin, M., Qiao, X., Cheng, Y., Li, X., Zhang, H., and Wu, J., A new insight into the evolution and functional divergence of SWEET transporters in Chinese white pear (Pyrus bretschneideri), Plant Cell Physiol., 2017, vol. 58, p. 839.
Wu, Y.W., Wang, Y.P., Shan, Y.X., and Qin, Q.P., Characterization of SWEET, family members from loquat and their responses to exogenous induction, Tree Genet. Genomes, 2017, vol. 13, p. 123.
Qin, Q., Cui, Y., Zhang, L., Lin, F., and Lai, Q., Isolation and induced expression of a fructokinase gene from loquat, Russ. J. Plant Physiol., 2014, vol. 61, p. 315.
Tamura, K., Stecher, G., Peterson, D., Filipski, A., and Kumar, S., MEGA6: Molecular Evolutionary Genetics Analysis version 6.0, Mol. Biol. Evol., 2013, vol. 30, p. 2725.
Li, H., Li, X., Xuan, Y., Jiang, J., Wei, Y., and Piao, Z., Genome wide identification and expression profiling of SWEET genes family reveals its role during Plasmodioph-ora brassicae-induced formation of clubroot in Brassica rapa,Front. Plant Sci., 2018, vol. 9, p. 207.
Wang, J., Yan, C., Li, Y., Hirata, K., Yamamoto, M., Yan, N., and Hu, Q., Crystal structure of a bacterial homologue of SWEET transporters, Cell Res., 2014, vol. 24, p. 1486.
Yang, G.X., Xu, H.F., Zhang, J., Wang, N., Fang, HC., Zou, Q., Wang, Y.C., Jiang, S.H., and Chen, X.S., Functional identification of a sugar transporter gene MdSWEET17 in apple, J. Plant Physiol., 2018, vol. 54, p. 1737.
Wang, L., Yao, L.N., Hao, X.Y., Li, N.N., Qian, W.J., Yue, C., Ding, C.Q., Zeng, J.M., Yang, Y.J., and Wang, X.C., Tea plant sweet transporters: expression profiling, sugar transport, and the involvement of CsSWEET16 in modifying cold tolerance in Arabidopsis, Plant Mol. Biol., 2018, vol. 96, p. 577.
Zhen, Q., Fang, T., Peng, Q., Liao, L., Zhao, L., Owiti, A., and Han, Y., Developing gene-tagged molecular markers for evaluation of genetic association of apple SWEET genes with fruit sugar accumulation, Hortic. Res., 2018, vol. 5, p. 14.
ACKNOWLEDGMENTS
We are grateful to Dr. Wenshun Hu, Fruit Research Institute, Fujian Academy of Agricultural Sciences and Dr. Junwei Chen, Institute of Horticulture, Zhejiang Academy of Agricultural Sciences for providing materials facilities and encouragement throughout.
Funding
This work was supported by National Natural Science Foundation of China (project no. 31570679), and the Science Technology Department of Zhejiang Province (project no. 2016C02052-3).
Author information
Authors and Affiliations
Contributions
J. H. Lu and L. Bai contributed equally to this work. Q.Q. and N.L. supervised the experiments and revised the manuscript. J.L. designed and performed the experiments, carried out data analysis and wrote the manuscript. L.B. revised the manuscript.
Corresponding authors
Ethics declarations
COMPLIANCE WITH ETHICAL STANDARDS
The authors declare that they have no conflict of interest. This article does not contain any studies involving animals or human participants as objects of research.
DATA ARCHIVING STATEMENT
The loquat SWEET17s identified in this study have been deposited with the GenBank Data Library (https://www. ncbi.nlm.nih.gov/genbank/) under accessions MK206393-MK206400.
Additional information
Abbreviations: GRAVY—grand average of hydropathy; MST—monosaccharide transporter gene; MW—molecular weight; pI—protein isoelectric point; SUT—sucrose transporter protein; SWEET—Sugars Will Eventually be Exported Transporters; TM—transmembrane structures; TMT—Tonoplast monosaccharide transporter; VGT—vacuolar glucose transporter.
Supplementary material
Rights and permissions
About this article
Cite this article
Lu, J.H., Bai, L., Qin, Q.P. et al. Isolation and Comparison of Eight SWEET17 Genes from Six Loquat Cultivars. Russ J Plant Physiol 67, 1063–1075 (2020). https://doi.org/10.1134/S1021443720060138
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1134/S1021443720060138