Living Within the Macrophage: Dimorphic Fungal Pathogen Intracellular Metabolism
- 1Department of Biology, Rhodes College, Memphis, TN, United States
- 2Department of Microbiology, Ohio State University, Columbus, OH, United States
Histoplasma and Paracoccidioides are related thermally dimorphic fungal pathogens that cause deadly mycoses (i.e., histoplasmosis and paracoccidioidomycosis, respectively) primarily in North, Central, and South America. Mammalian infection results from inhalation of conidia and their subsequent conversion into pathogenic yeasts. Macrophages in the lung are the first line of defense, but are generally unable to clear these fungi. Instead, Histoplasma and Paracoccidioides yeasts survive and proliferate within the phagosomal compartment of host macrophages. Growth within macrophages requires strategies for acquisition of sufficient nutrients (e.g., carbon, nitrogen, and essential trace elements and co-factors) from the nutrient-depleted phagosomal environment. We review the transcriptomic and recent functional genetic studies that are defining how these intracellular fungal pathogens tune their metabolism to the resources available in the macrophage phagosome. In addition, recent studies have shown that the nutritional state of the macrophage phagosome is not static, but changes upon activation of adaptive immune responses. Understanding the metabolic requirements of these dimorphic pathogens as they thrive within host cells can provide novel targets for therapeutic intervention.
Introduction
Histoplasma and Paracoccidioides species cause respiratory and systemic disease in mammals (histoplasmosis and paracoccidioidomycosis, respectively). Disease results from inhalation of aerosolized fungal conidia from the environmental mold form. Severity of disease ranges from subclinical to acute, and is largely a function of the dose inhaled and the immunological defenses of the host (Kauffman, 2009; Queiroz-Telles and Escuissato, 2011; Salzer et al., 2018). In immunocompromised individuals (e.g., HIV patients), infection can result in life-threatening disseminated disease. Recent phylogenetic analyses based on genome sequencing efforts have split the original Histoplasma capsulatum and Paracoccidiodies brasiliensis designations into additional species (Van Dyke et al., 2019). Although some physical and biochemical differences exist among these species groups, in this review we will refer to studies in each fungus by the respective genus name unless necessary to specifically highlight findings unique to a species.
The Histoplasma and Paracoccidiodies genera comprise closely related fungi which are characterized by thermal dimorphism. Conidia, produced by the environmental mycelial forms, convert into pathogenic yeast forms in response to the elevated temperature following inhalation by a mammalian host. The yeasts are budding yeasts, with Paracoccidioides yeasts typically characterized by multiple budding daughter cells around a central yeast body. A hallmark of these dimorphic fungal pathogens is the link between pathogenesis and conversion into yeasts (Medoff et al., 1987; Maresca and Kobayashi, 1989; Aristizabal et al., 1998; Borges-Walmsley et al., 2002).
During infection, Histoplasma and Paracoccidioides yeast cells are phagocytosed by cells of the innate immune system. Studies with cultured phagocytes (i.e., macrophages) show roughly 10%-30% of Paracoccidioides yeasts are taken up by macrophages (Soares et al., 2010; da Silva et al., 2016) while up to 60% of Histoplasma yeasts are phagocytosed (Newman et al., 1990; Garfoot et al., 2016) within 60–90 min in tissue culture conditions. Studies of respiratory infection in mice show that nearly all Histoplasma yeasts are found within CD45+ phagocytes (Deepe et al., 2008). Thus, Histoplasma yeasts reside almost exclusively within phagocytes during infection in vivo, and many, but not all Paracoccidioides yeasts are intracellular. Non-activated host phagocytes are relatively ineffective in killing and controlling Histoplasma and Paracoccidioides yeasts. Activation of phagocytes during adaptive immunity by T helper cell type 1 (Th1) is typically required for host control of these fungi (Allendoerfer and Deepe, 1997, 1998; Souto et al., 2000; Calich et al., 2008; de Castro et al., 2013) thereby distinguishing Histoplasma and Paracoccidioides from opportunistic fungal pathogens, which are readily controlled without T-cell activation. Recently, IL-17 signaling was shown to also contribute to the control of Paracoccidioides (Tristão et al., 2017).
To effectively use phagocytes as host cells, Histoplasma and Paracoccidioides yeasts express mechanisms that facilitate binding to host phagocytes and survival of macrophage defenses. Although the fungal cell wall contains β-glucans which can alert the immune system to the presence of a fungal invader, Histoplasma yeasts effectively hide this β-glucan signature by overlaying the immunostimulatory β-glucans with α-glucan polysaccharides (Rappleye et al., 2004, 2007). In addition, Histoplasma yeasts secrete an endoglucanase (Eng1) that trims away remaining exposed β-glucans (Garfoot et al., 2016). Although a similar mechanism has not been shown for Paracoccidioides, the cell wall of Paracoccidioides yeasts also has a high content of α-glucan (Kanetsuna et al., 1972) and decreased α-glucan is associated with decreased virulence (San-Blas et al., 1977). Histoplasma expresses two extracellular antioxidant enzymes, Sod3 and CatB, which together effectively eliminate phagocyte-derived reactive oxygen (Youseff et al., 2012; Holbrook et al., 2013) and facilitate Histoplasma survival during uptake by macrophages. Consistent with the pathogenesis-enabling role of these mechanisms, α-glucan production and secretion of Eng1, Sod3, and CatB characterize the virulent yeasts, but not the avirulent mycelial cells.
Pathogenic yeast not only survive the encounter with phagocytes, but also use the phagocyte as a permissive niche during infection. As long-term intracellular residents of the macrophage, Histoplasma and Paracoccidioides yeasts must adapt their metabolism to nutrients available in the phagosome. The phagosome environment is generally assumed to be nutrient poor, yet these fungi are able to scavenge sufficient carbon, nitrogen, sulfur, and micronutrients from the host cell to support fungal growth and replication. Many studies have used gene expression profiling to infer which metabolic pathways are active during residence within the phagosome. Interpretation of differentially expressed genes, however, is complicated by the in vitro growth condition used as the comparison. Furthermore, a large number of genes that characterize the pathogenic yeast phase of the dimorphic fungi are controlled by phase-differentiation factors such as the Ryp proteins (Shen and Rappleye, 2017; Beyhan and Sil, 2019) suggesting that some metabolic genes may not be regulated by the nutritional environment, but instead their expression is set by the differentiation state (i.e., yeasts vs. mycelia). Genetic studies, and the functional tests it facilitates, are a means to more conclusively determine the metabolism used by intracellular yeasts to parasitize the macrophage. Recent studies highlighted below integrate key findings that define the metabolic pathways required for intracellular yeast proliferation, and by extension, the host substrates that are consumed by these fungal pathogens during infection.
Intraphagosomal Carbon Metabolism
Glycolysis
Most in vitro fungal growth media is based on glucose as the primary carbon source since glucose affords rapid fungal growth. Although this rich carbon source may reflect some host environments (e.g., the bloodstream), other host niches, including the macrophage phagosome are characterized by alternative carbon sources. Gene expression studies in which specific metabolic pathways are up-regulated in specific host environments can reveal the presence of different carbon sources. Bailão et al. (2006) conducted the first study with Paracoccidioides yeasts to determine which genes are up-regulated during infection. Comparing the gene expression profile of yeasts grown in human blood to yeasts grown in vitro, Bailão et al. (2006) found that glyceraldehyde 3-phosphate dehydrogenase (GAPDH) was up-regulated in blood. Since GAPDH is involved in both glycolysis and gluconeogenesis, it remains unclear if glucose catabolism contributes to Paracoccidioides infection of host cells. While some Paracoccidioides cells may be taken up by phagocytes in blood (e.g., neutrophils), significant numbers may remain extracellular. Thus, the results might reflect the transcriptional responses of a mixed population consisting of both extracellular yeasts and intracellular yeasts from different types of phagocytes. Thus, the high concentration of extracellular glucose in the blood, not the intracellular phagosome environment, may be the primary driver of GAPDH up-regulation.
Profiling of yeasts resident within macrophages offers a more direct assessment of the transcriptional profiles supporting intraphagosomal growth. Tavares et al. (2007) conducted a differential expression study comparing the gene expression profile of Paracoccidioides yeasts grown in murine peritoneal macrophages and those grown in vitro in rich media. Using a transcriptional microarray, the authors found that the gene encoding the glycolysis specific enzyme phosphofructokinase (PFK1) was down-regulated in intracellular Paracoccidioides yeasts. A separate transcriptional study that profiled Paracoccidioides yeasts from the lung identified a sugar transporter that was down-regulated relative to yeasts grown in vitro in a glucose-rich medium [2% glucose (Lacerda Pigosso et al., 2017)]. This suggests that glucose catabolism is not a primary metabolic pathway for Paracoccidioides intracellular growth. However, in the Tavares study (Tavares et al., 2007), RT-PCR data was unable to confirm the microarray results. Proteomic analysis of Paracoccidioides intracellular yeasts found that hexokinase (Hxk1) was down-regulated compared to yeast grown in a glucose-rich medium in vitro consistent with decreased metabolism of sugars (Parente-Rocha et al., 2015). The transcriptional signature of intracellular Histoplasma yeasts also shows down-regulation of a glycolytic response when growing within macrophages (Shen et al., 2020). As with Paracoccidioides, PFK1 was down-regulated in Histoplasma yeasts. Genes encoding other glucose-catabolism factors including pyruvate kinase (PYK1) and the hexose/glucose kinases (HXK1 and GLK1) were also down-regulated. The finding that multiple components of glycolysis showed decreased expression provides stronger support for the metabolism of alternative carbon sources during intracellular proliferation. As an independent indicator of the lack of hexose catabolism by intracellular Histoplasma yeasts, metabolic assays (i.e., Seahorse assays) of Histoplasma yeasts isolated from macrophages showed the yeasts were glycolytically inactive (Shen et al., 2020) consistent with the transcriptional profiles. Definitive evidence of the lack of hexose catabolism by intracellular yeasts came from the analysis of glycolysis-deficient Histoplasma yeasts. Simultaneous depletion of Histoplasma's only hexose/glucose kinases (Hxk1 and Glk1) impaired the ability of Histoplasma yeast to grow on glucose as the carbon source in vitro, but Hxk1 and Glk1 loss did not prevent Histoplasma yeast growth within macrophages (Shen et al., 2020). Most revealing, the depletion of Hxk1 and Glk1 and no effect on Histoplasma respiratory infection in vivo (Shen et al., 2020). These data demonstrate that Histoplasma yeasts, and likely also Paracoccidioides yeasts, do not catabolize hexoses to grow within macrophages. As both Histoplasma and Paracoccidioides efficiently use glucose in vitro, this suggests that it is unlikely that hexoses are available to yeasts within the macrophage phagosome.
Gluconeogenesis
The lack of hexose utilization suggests that gluconeogenic substrates are likely metabolized by intracellular yeasts to provide energy and essential biomolecules including glucans for the polysaccharide-rich cell wall. A combined transcriptomic and proteomics study compared Paracoccidioides yeasts isolated from mouse lungs after 6 h of infection to yeasts grown in a glucose- and peptide-rich medium in vitro [Brain Heart Infusion medium (Lacerda Pigosso et al., 2017)]. The PCK1 gene, encoding phosphoenolpyruvate carboxykinase which catalyzes the first committed step of gluconeogenesis, was transcriptionally up-regulated in vivo, suggesting the yeasts resided in a glucose-depleted environment. However, the proteomics analysis failed to detect Pck1 at the protein level in yeasts isolated from the lung. As only a portion of Paracoccidioides yeasts get internalized by phagocytes, it is unclear whether the up-regulation of PCK1 reflects the intracellular or extracellular nutritional environment during in vivo infection and whether 6 h of infection is sufficient for transcriptional or proteomic adaptive responses. One of the limitations of this study was the recovery of low numbers of yeasts which could impact the sensitivity of their analyses. Many normally-abundant proteins were not detected in the proteomic data and key enzymes involved in the central carbon metabolism (i.e., pyruvate kinase, citrate synthase, and isocitrate dehydrogenase) that should be abundant in yeast cells, regardless of growth condition, were not recovered from the proteome data. No conclusions can be made from the absence of data, but an independent study of intramacrophage Paracoccidioides yeasts also showed up-regulation of the PCK1 gene after 9 h of infection (Derengowski et al., 2008). In addition, Parente-Rocha et al. (2015) found that gluconeogenesis-specific enzyme fructose-1,6-bisphosphatase was modestly increased (1.5-fold) in Paracoccidioides yeasts recovered from macrophages, providing further support of gluconeogenesis-based metabolism in intracellular yeasts.
Studies of Histoplasma yeasts isolated from macrophages 24 h after infection shows PCK1 expression is up-regulated in intracellular yeasts (Shen et al., 2020). Elimination of Pck1 activity prevents growth of Histoplasma yeasts within macrophages but not in vitro growth in glucose-containing medium confirming that the phagosome environment lacks hexose substrates (Shen et al., 2020). Histoplasma yeasts lacking Pck1 function are severely attenuated in a murine model of respiratory infection as early as 2 days post-infection and this growth defect in vivo persisted throughout the entire 8-day infection period, indicating that the Histoplasma-containing phagosome remains depleted of hexose substrates. The gluconeogenesis-specific fructose-1,6-bisphosphatase (Fbp1) is also required for Histoplasma yeast virulence in macrophages and in mice (Shen et al., 2020). These findings are consistent with the results that glycolytic enzymes are not required for Histoplasma full virulence in vivo. Collectively, these findings indicate that Histoplasma yeasts rely on metabolizing gluconeogenic carbon sources to proliferate within macrophages (Figure 1) and this metabolism likely characterizes intracellular Paracoccidioides yeasts.

Figure 1. Intramacrophage carbon metabolism of Histoplasma and Paracoccidioides. Schematic map depicts the central metabolic pathways for the metabolism of different carbon-containing molecules with key carbon intermediates shown. Enzymes catalyzing critical steps of glycolysis (Hxk1/Glk1, Pfk1, and Pyk1), gluconeogenesis (Pck1 and Fbp1), and fatty acid utilization (Fox1 for β-oxidation of fatty acids, and Icl1 and Mls1 for the glyoxylate cycle for incorporation of acetyl-CoA into biomolecules) are indicated. Names of enzymes are colored based on the down-regulation (magenta text) or up-regulation (blue text) in fungal cells within macrophages. in situations where expression data is conflicting, text is colored orange. For expression data that is discrepant between Paracoccidioides and Histoplasma, the respective genes are provided by a species prefix (Pb and Hc, respectively). Arrows indicate the flow of carbon through metabolic pathways. Enzymatic reactions that are not required for intracellular growth are indicated with magenta arrows. Reactions necessary for proliferation in the phagosome and for infection in vivo are indicated with blue arrows. Points of entry for amino acids as potential carbon substrates are indicated in yellow. Enzyme abbreviations: Hxk1, hexokinase; Glk1, glucokinase; Pfk1, phosphofructokinase; Pyk1, pyruvate kinase; Pck1, phosphoenolpyruvate carboxykinase; Fbp1, fructose 1,6-bisphosphatase; Fox1, fatty acyl-CoA oxidase; Icl1, isocitrate lyase. Metabolite abbreviations: G6P, glucose 6-phosphate; F6P, fructose 6-phosphate; F1, 6BP, fructose 1,6-bisphosphate; GAP, glyceraldehyde 3-phosphate; PEP, phosphoenolpyruvate; CIT, citrate; α-KG, α-ketoglutarate; SUC, succinate; OXA, oxaloacetate.
Fatty Acids Utilization
What are the gluconeogenic substrates derived from the host that support yeast proliferation within phagosomes? One potential source is host fatty acids. These long-chain carbon molecules can be metabolized to acetyl-CoA (β-oxidation) which subsequently enters the TCA cycle to generate energy or can be assimilated into carbon biomass through the glyoxylate shunt. The up-regulation of the glyoxylate shunt can indicate utilization of fatty acids as the carbon source. In one study (Derengowski et al., 2008), the expression of key genes involved in glyoxylate shunt [isocitrate lyase (ICL1) and malate synthase (MLS1)] in Paracoccidioides yeasts was measured during infection of J774 macrophages using semi-quantitative RT-PCR. Expression of ICL1 increased ~3-fold in macrophages compared to growth in Fava-Neto medium (a rich medium containing high glucose, peptides, and amino acids). However, expression of MLS1 showed only a minor increase in expression. Isocitrate lyase protein (Icl1) increased 1.6-fold in Paracoccidioides yeasts recovered from J774 macrophages (Parente-Rocha et al., 2015). The up-regulation of ICL1 is consistent with results profiling Candida albicans and Cryptococcus neoformans yeasts when interacting with macrophages (Lorenz et al., 2004; Fan et al., 2005). However, Tavares et al. (2007) reported conflicting results that ICL1 was not up-regulated in Paracoccidioides yeasts within peritoneal macrophages, possibly due to in vitro growth conditions used for comparison. Expression profiling of Paracoccidioides yeasts 6 h after infection of mice also found an indication of the up-regulation of the glyoxylate shunt, but only the MLS1 gene, not ICL1 (Lacerda Pigosso et al., 2017). Growth of Paracoccidioides yeasts on acetate in vitro increases both ICL1 and MSL1 expression by a similar magnitude (Derengowski et al., 2008), suggesting the one-sided increases in ICL1 or MLS1 expression by Paracoccidioides yeasts in macrophages or in vivo do not indicate simple induction of the glyoxylate cycle. These inconsistencies complicate inferences about the role of the glyoxylate cycle in intracellular Paracoccidioides yeasts.
As with the glyoxylate cycle, indications of fatty acid catabolism by Paracoccidioides yeasts are also variable. Enzymes catalyzing the breakdown of fatty acids via β-oxidation have been examined for Paracoccidioides yeasts both in macrophages and in vivo. While enoyl-CoA hydratase (which catalyzes the second step in fatty acid metabolism) was up-regulated in Paracoccidioides yeasts from J774 macrophages, 3-keyoacyl-CoA thiolase (which catalyzes the last step liberating acetyl-CoA) was down-regulated in these same intracellular yeasts (Parente-Rocha et al., 2015). Transcriptional profiling of Paracoccidioides yeast showed three different genes annotated as encoding acyl-CoA dehydrogenases, which catalyze the first step in fatty acid breakdown, were increased during respiratory infection of mice (Lacerda Pigosso et al., 2017). One of these was increased 13-fold over yeast grown in vitro. However, Parente-Rocha et al. (2015) did not detect any increased acyl-CoA dehydrogenases in Paracoccidioides yeasts resident within cultured macrophages. These inconsistencies as well as the potential profiling of a mixed population of intra- and extracellular Paracoccidioides yeasts following infection of mice leave the role of fatty acid metabolism unclear for Paracoccidioides.
In contrast, Histoplasma yeasts within macrophages do not present hallmarks of fatty acid utilization. The transcription of genes encoding enzymes of the glyoxylate cycle (ICL1 and MLS1) as well as the first step in fatty acid β-oxidation [fatty acyl-CoA oxidase (FOX1)] were all down-regulated in intracellular Histoplasma yeasts (Shen et al., 2020). Histoplasma yeasts are unable to metabolize exogenous fatty acids as the carbon source in vitro. Thus, changes in β-oxidation enzymes may reflect modulation of endogenous fatty acids unrelated to host carbon substrates. The most conclusive evidence of the lack of host fatty acid consumption by Histoplasma yeasts was obtained by functional studies with yeast strains depleted of key enzymes in fatty acid utilization. Histoplasma yeasts lacking Icl1 or lacking Fox1 are fully virulent in cultured macrophage infection as well as in respiratory infection of mice (Shen et al., 2020). The lack of any requirement for fatty acid utilization by intracellular Histoplasma yeasts differs from that of other fungal pathogens [e.g., C. albicans (Lorenz and Fink, 2001) and C. neoformans (Kretschmer et al., 2012)], which occupy diverse extracellular niches or are only transiently found within phagocytes. As Histoplasma yeasts are nearly all within phagocytes during infection, this suggests that Histoplasma yeasts have adapted to a more prolonged residence within the phagosome and do not utilize host fatty acids as a major carbon source, either because exogenous fatty acids are unavailable within the phagosome or because other carbon sources are more consistently obtained in this intracellular compartment. Whether intracellular Paracoccidioides yeast metabolism mirrors that of intracellular Histoplasma yeasts or if it follows the paradigm of transient occupants of the phagocyte remains to be determined.
Amino Acid Catabolism
The above studies indicate that both Paracoccidioides and Histoplasma yeasts exploit gluconeogenic substrates for intracellular proliferation. What are these alternative host carbon substrates obtained from the host macrophage, and specifically which are found within the phagosome? Carbon source utilization tests in vitro showed that the spectrum of carbon sources which can be metabolized by yeasts differs from those that are metabolized by Histoplasma mycelia, suggesting that the pathogenic yeasts are more metabolically streamlined (Shen et al., 2020). For example, Histoplasma yeasts can metabolize hexoses and hexosamines but not pentoses or disaccharides, some of which are metabolized by mycelia. In addition, Histoplasma yeasts can use a mix of amino acids as a non-carbohydrate carbon source. Given hexose-catabolism and fatty acids catabolism pathways are not required for Histoplasma intracellular proliferation, this focuses attention on amino acids as potential sources of carbon for pathogenic-phase yeasts within the phagosome. Indeed, the phagosome/phagolysosome is a degradative organelle harboring numerous proteases that could generate amino acids and short peptides through host protein degradation.
Expression of genes involved in amino acid biosynthesis hints to the absence of certain amino acids within the phagosome. Bailão et al. (2006) showed that the gene catalyzing the last step of glutamine synthesis (i.e., GLN1) in Paracoccidioides is up-regulated in fresh human blood, suggesting that glutamine in the human blood environment is not sufficient to support Paracoccidioides growth. However, the intracellular residence of Paracoccidioides is unknown in this study. In peritoneal macrophages, Paracoccidioides yeasts up-regulated the gene involved in methionine synthesis (METG) suggesting that methionine is not available within macrophages (Tavares et al., 2007). Differential gene expression of Paracoccidioides from respiratory infection showed genes encoding enzymes involved in the metabolism of glycine, serine, threonine, methionine, and lysine are down-regulated (Lacerda Pigosso et al., 2017). Also, down-regulated were genes encoding 4-hydroxyphenylpyruvate dioxygenase (HPD1) and homogentisate dioxygenase (HGD1), which are involved in phenylalanine and tyrosine degradation suggesting these aromatic amino acids are not consumed from the host. On the other hand, Parente-Rocha's study of Paracoccidioides yeasts from macrophages showed increased abundance of Hpd1 (Parente-Rocha et al., 2015) contradicting the in vivo gene expression study. Expression studies thus do not provide definitive answers, especially considering the interconversion of many amino acids. Functional evidence comes from a study that used the chemical compound CP1, which is known to inhibit chorismate synthase, the key enzyme in biosynthesis of aromatic amino acids (i.e., tyrosine, tryptophan, and phenylalanine). Treatment of Paracoccidioides-infected mice with CP1 reduced fungal burdens ~10-fold compared to untreated mice (Rodrigues-Vendramini et al., 2019) suggesting that Paracoccidioides yeasts must synthesize at least one of the three to make up for the deficiency in the environment in which Paracoccidioides is found during lung infection.
Histoplasma yeasts can catabolize amino acids as the sole carbon source demonstrating the potential to use host amino acids within the phagosome. On the other hand, Histoplasma yeasts cannot utilize model host protein substrates for carbon (e.g., bovine serum albumin, gelatin, and hemoglobin). Prior digestion of these proteins with proteinases, including the phagosomal proteinase Cathepsin D, renders the proteins to a state that can support the growth of Histoplasma yeasts (Shen et al., 2020). Thus, it appears that Histoplasma yeasts can take up amino acids and select peptides to use as carbon but does not metabolize intact proteins. This would be consistent with residence within the phagosome where host proteinases may supply the degradative capacity to liberate amino acids for consumption by intraphagosomal Histoplasma yeasts. Indeed, the expression of many metabolic genes of Histoplasma yeasts within the macrophage is highly similar to the profile of Histoplasma yeasts growing on amino acids in vitro (Shen et al., 2020). The expression of various peptidases are increased in Paracoccidioides yeast within J774 macrophages (Parente-Rocha et al., 2015) and Paracoccidioides yeast secrete a serine proteinase during infection (Lacerda Pigosso et al., 2017), suggesting that proteins and peptides are hydrolyzed to amino acids before consumption. Nonetheless, not all amino acids are sufficiently present in the phagosome to support fungal growth. A Histoplasma alanine auxotroph showed impaired virulence in both macrophages and in vivo, indicating alanine is deficient in the phagosomal environment (Shen et al., 2020). Combined with the lack of hexose and fatty acid catabolism described for Histoplasma yeasts above, it is likely that Histoplasma yeasts within the phagosome catabolize amino acids derived from the host which are then processed through gluconeogenic pathways to supply carbohydrates for biomass (Figure 1). Further investigations will be needed to define which amino acids are available within the phagosome and which are catabolized by the intracellular yeasts.
Intraphagosomal Nitrogen and Sulfur
One of the advantages of amino acid catabolism within the phagosome is that amino acids provide yeasts not only carbon, but also a source of nitrogen and potentially sulfur. Consistent with amino acids providing nitrogen to intracellular yeasts, Paracoccidioides yeasts recovered from macrophages have increased abundance of various aminotransferases (Parente-Rocha et al., 2015). However, in Paracoccidioides yeasts from lung infections, a branched-chain amino acid aminotransferase is down-regulated again showing inconsistencies between the lung and the macrophage models. The transcriptome of yeasts from lungs showed increased expression of amino acid permeases and peptide transporters (Lacerda Pigosso et al., 2017) consistent with yeasts importing host amino acids and peptides during infection. Differential gene expression analysis of Histoplasma yeasts compared to mycelia also show three genes encoding aminotransferases are up-regulated in yeasts (Edwards et al., 2013). Interestingly, studies of Paracoccidioides yeasts consistently highlight formamidase as one of the most up-regulated genes or proteins increased in expression during infection of mice or cultured macrophages (Parente-Rocha et al., 2015; Lacerda Pigosso et al., 2017). This suggests formamidase is a central step in nitrogen acquisition and metabolism for Paracoccidioides yeast during infection. Alternatively, formamidase may be particularly repressed in the conditions used for in vitro growth used as comparison. While the in vivo substrate(s) for formamidase is/are unknown (e.g., formamide), arylformamidases are involved in tryptophan degradation which may signal utilization of tryptophan during infection.
Cysteine and methionine represent sulfur-containing amino acids that can provide organic sulfur to fungal pathogens. Changes in cysteine metabolism during transitions between Histoplasma yeast and mycelial phases are well-known (Stetler and Boguslawski, 1979; Maresca et al., 1981). Despite a pathway for inorganic sulfur assimilation (e.g., sulfate) that is operative in Histoplasma mycelia, Histoplasma yeasts require an organic sulfur source which can be supplied by cysteine (Salvin, 1949). Comparison of Histoplasma yeast and mycelial transcriptomes showed increased expression of a permease for methionine and cysteine (Hwang et al., 2003), suggesting the pathogenic yeast state is primed to uptake exogenous sulfur-containing amino acids. Despite the auxotrophy of Histoplasma yeasts for organic sulfur, Histoplasma is fully virulent indicating that organic sulfur is available to Histoplasma yeasts within the phagosome. While the ultimate source of organic sulfur for intracellular fungal growth remains unknown, cysteine is a logical candidate, which may be derived from protein degradation or even hydrolysis of abundant glutathione in the macrophage.
Acquisition of Micronutrients
Essential Vitamins
While required in smaller quantities than carbon or nitrogen, vitamins are essential micronutrients for growth. This nutritional requirement can be met either by vitamin acquisition from the host or through de novo synthesis by fungal cells. Biosynthesis of vitamins by Paracoccidioides and Histoplasma appears to be the primary source for these essential co-factors. Paracoccidioides yeasts infecting the murine lung up-regulate the gene involved in thiamine biosynthesis (THI13), suggesting that thiamine from the host is not available to yeasts during infection (Lacerda Pigosso et al., 2017). Parente-Rocha et al. (2015) similarly found that Paracoccidioides yeasts within macrophages have increased expression of enzymes involved in the synthesis of thiamine as well as pyridoxine and riboflavin. Although increased expression of these enzymes suggests yeasts synthesize vitamins from simpler metabolic intermediates during infection, expression was measured relative to yeast growth in vitro in a vitamin-rich medium. More direct evidence for the lack of vitamins within the macrophage host comes from functional studies in Histoplasma. A forward genetics screen for mutants unable to proliferate within macrophages isolated a mutant with a disruption of the RIB2 gene, which encodes a deaminase catalyzing the third step of riboflavin biosynthesis (Garfoot et al., 2014). While Histoplasma yeast lacking Rib2 function could infect macrophages, they were severely attenuated for growth in macrophages as well as attenuated in lung infection. This indicates that Histoplasma cannot acquire sufficient exogenous riboflavin from the phagosomal environment to support its intracellular growth. In addition, depletion of the pantothenate biosynthesis enzyme, Pan6, impaired Histoplasma virulence in mice indicating that pantothenate is also scarce in the phagosomal environment (Garfoot et al., 2014). On the other hand, depletion of biotin biosynthesis did not affect Histoplasma virulence. Although this may suggest that host biotin can be scavenged within the phagosome, it is also possible that the Histoplasma yeast had stored sufficient biotin from the pre-growth. In support of this, depletion of biotin synthase (Bio2) did not impair growth in vitro until yeasts were passaged multiple times in biotin-deficient media. Thus, the macrophage phagosome compartment in general lacks essential vitamins thereby forcing intracellular yeasts to rely on de novo synthesis to meet this nutritional requirement.
Trace Metals
In addition to vitamin co-factors, trace metals are essential for yeast growth and are variably available within the phagosome depending on the activation state of macrophages. Iron acquisition is essential for Histoplasma and Paracoccidioides virulence. Attenuation of Paracoccidioides and Histoplasma yeast growth in macrophages by chloroquine, which impairs phagosome acidification and availability of iron, is reversed by supplementation with exogenous iron (Newman et al., 1994; Dias-Melicio et al., 2006). In addition, supplementation of exogenous iron exacerbated Paracoccidioides infection in mice (Parente et al., 2011). Iron restriction of Paracoccidioides yeasts in vitro induces genes encoding enzymes involved in siderophore biosynthesis and transport (Parente et al., 2011; Silva-Bailão et al., 2014). However, induction of siderophore biosynthesis and transport was not observed in Paracoccidioides yeasts isolated from murine macrophages or in vivo. Among different iron acquisition mechanisms, Histoplasma yeasts secrete siderophores for scavenging rare iron (Hwang et al., 2008; Hilty et al., 2011). Genes encoding siderophore synthesis enzymes are located in a genetic cluster which is activated under low iron conditions through the action of Sre1, a GATA-family transcription factor (Hwang et al., 2008). Loss of siderophore biosynthesis impairs intracellular Histoplasma growth indicating the phagosome is a low iron compartment. However, studies of Histoplasma infection of mice indicate that siderophores are not required for proliferation of Histoplasma pathogenesis until 2-weeks post-infection which coincides with the adaptive immunity stage (Hwang et al., 2008). These results indicate that during early infection, sufficient iron is found within the macrophage phagosome, but after activation of host cells, iron becomes limiting for fungal growth.
Similar to iron, zinc levels in the phagosome can change depending on the activation state of macrophages. Initially, zinc is available for intracellular Histoplasma yeast growth, but becomes limiting as macrophages are activated by the GM-CSF cytokine to produce zinc-sequestering metallothioneines (Subramanian Vignesh et al., 2013). Histoplasma yeasts produce the Zrt2 zinc transporter to acquire zinc from the environment and this transporter is required for Histoplasma virulence in mice, but only after 3 days of infection (Dade et al., 2016). These data indicate that host macrophages can use restriction of trace metals from the phagosome to combat intracellular proliferation of yeasts, particularly after macrophage activation.
Host macrophages can use both abundance and restriction of available copper to curtail intracellular pathogen growth. Tavares et al. (2007) found that Paracoccidioides yeasts down-regulate a high-affinity copper transporter by 17-fold within cultured macrophages after 6 h, suggesting that the Paracoccidioides-containing phagosome has sufficient copper to support Paracoccidioides growth. This same down-regulation of the copper transporter was also observed during lung infection of mice at 6 h (Lacerda Pigosso et al., 2017) suggesting the lung environment has ample copper during early infection. The Histoplasma genome encodes a homologous copper transporter (Ctr3) whose transcription is induced at low copper concentrations (<150 nM) (Shen et al., 2018). Through use of a transcriptional fusion of the copper-repressible CTR3 promoter with a fluorescent protein, available copper levels within the Histoplasma-containing phagosome could be estimated. In resting macrophages, including alveolar macrophages which Histoplasma cells initially encounter during infection, phagosomal copper levels are roughly 300 nM. At this concentration, Histoplasma yeasts can acquire sufficient copper without needing the Ctr3 transporter. Upon activation of macrophages with IFN-γ, the available copper in the phagosome significantly decreases resulting in up-regulation of the Ctr3 transporter (Shen et al., 2018). Consistent with the phagosomal copper levels inferred from transcription studies, the Ctr3 transporter is not required for Histoplasma proliferation within resting macrophages (i.e., higher phagosomal copper levels) but becomes necessary following macrophage activation as phagosomal copper becomes restricted (Figure 2; Shen et al., 2018). These results indicate that copper availability in the macrophage phagosome, as well as that of iron and zinc, is dynamic and that activation of macrophages imposes a metal restriction strategy to impair the intracellular proliferation of yeasts.
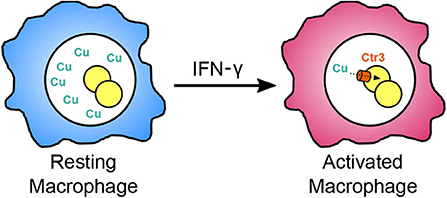
Figure 2. Restriction of available copper within the phagosome following macrophage activation. During initial infection, Histoplasma infects resting macrophages (blue), and resides within a phagosomal compartment with sufficiently available copper (green Cu) for fungal proliferation without requiring the high affinity copper transporter Ctr3. Upon activation of macrophages by IFN-γ during adaptive immunity (magenta), copper in the phagosome becomes restricted forcing Histoplasma yeast to rely on Ctr3 to import copper.
Conclusion
Initial studies on fungal pathogens often focused on virulence mechanisms for host cell infection and survival. The studies highlighted above indicate that mechanisms of fungal metabolism are an integral component of intracellular fungal pathogenesis. While in vitro culture of fungi has typically used glucose- and peptone-rich media, the study of long-term phagosomal residents like Histoplasma and Paracoccidioides yeasts is revealing that alternate substrates, not hexoses, are available within the phagosome compartment. Furthermore, these dimorphic fungi have tuned their metabolism to exploit these resources during yeast infection of host cells. Consequently, media used for in vitro propagation of the yeasts should be re-evaluated for their physiological relevance to the nutritional conditions actually experienced by the yeast cells. The more-specialized metabolism used by yeasts within the phagosome creates opportunities for anti-fungal interventions. The finding that some micronutrient levels in the phagosome can change following macrophage activation and subsequent control of intracellular fungal proliferation further underscores the potential of targeting metabolic pathways for therapeutic control of these primary pathogens.
Author Contributions
QS and CR conceived of the review topic, performed the literature analysis, and composed the review article. All authors contributed to the article and approved the submitted version.
Funding
Research by QS and CR was supported by the National Institute of Allergy and Infectious Diseases of the National Institutes of Health under award numbers R21-AI137714 and R01-AI148561. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
References
Allendoerfer, R., and Deepe, G. S. (1997). Intrapulmonary response to Histoplasma capsulatum in gamma interferon knockout mice. Infect. Immun. 65, 2564-2569.
Allendoerfer, R., and Deepe, G. S. (1998). Blockade of endogenous TNF-alpha exacerbates primary and secondary pulmonary histoplasmosis by differential mechanisms. J. Immunol. 160, 6072–6082.
Aristizabal, B. H., Clemons, K. V., Stevens, D. A., and Restrepo, A. (1998). Morphological transition of Paracoccidioides brasiliensis conidia to yeast cells: in vivo inhibition in females. Infect. Immun. 66, 5587–5591. doi: 10.1128/IAI.66.11.5587-5591.1998
Bailão, A. M., Schrank, A., Borges, C. L., Dutra, V., Walquíria Inês Molinari-Madlum, E. E., Soares Felipe, M. S., et al. (2006). Differential gene expression by Paracoccidioides brasiliensis in host interaction conditions: representational difference analysis identifies candidate genes associated with fungal pathogenesis. Microbes Infect. 8, 2686–2697. doi: 10.1016/j.micinf.2006.07.019
Beyhan, S., and Sil, A. (2019). Sensing the heat and the host: virulence determinants of Histoplasma capsulatum. Virulence 10, 793–800. doi: 10.1080/21505594.2019.1663596
Borges-Walmsley, M. I., Chen, D., Shu, X., and Walmsley, A. R. (2002). The pathobiology of Paracoccidioides brasiliensis. Trends Microbiol. 10, 80–87. doi: 10.1016/S0966-842X(01)02292-2
Calich, V. L. G., da Costa, T. A., Felonato, M., Arruda, C., Bernardino, S., Loures, F. V., et al. (2008). Innate immunity to Paracoccidioides brasiliensis infection. Mycopathologia 165, 223–236. doi: 10.1007/s11046-007-9048-1
da Silva, T. A., Roque-Barreira, M. C., Casadevall, A., and Almeida, F. (2016). Extracellular vesicles from Paracoccidioides brasiliensis induced M1 polarization in vitro. Sci Rep 6:35867. doi: 10.1038/srep35867
Dade, J., DuBois, J. C., Pasula, R., Donnell, A. M., Caruso, J. A., Smulian, A. G., et al. (2016). HcZrt2, a zinc responsive gene, is indispensable for the survival of Histoplasma capsulatum in vivo. Med. Mycol. 54, 865–875. doi: 10.1093/mmy/myw045
de Castro, L. F., Ferreira, M. C., da Silva, R. M., Blotta, M. H., Longhi, L. N., and Mamoni, R. L. (2013). Characterization of the immune response in human paracoccidioidomycosis. J. Infect. 67, 470–485. doi: 10.1016/j.jinf.2013.07.019
Deepe, G. S., Gibbons, R. S., and Smulian, A. G. (2008). Histoplasma capsulatum manifests preferential invasion of phagocytic subpopulations in murine lungs. J. Leukoc. Biol. 84, 669–678. doi: 10.1189/jlb.0308154
Derengowski, L. S., Tavares, A. H., Silva, S., Procópio, L. S., Felipe, M. S. S., and Silva-Pereira, I. (2008). Upregulation of glyoxylate cycle genes upon Paracoccidioides brasiliensis internalization by murine macrophages and in vitro nutritional stress condition. Med. Mycol. 46, 125–134. doi: 10.1080/13693780701670509
Dias-Melicio, L. A., Moreira, A. P., Calvi, S. A., and Soares, A. M. V. (2006). Chloroquine inhibits Paracoccidioides brasiliensis survival within human monocytes by limiting the availability of intracellular iron. Microbiol. Immunol. 50, 307–314. doi: 10.1111/j.1348-0421.2006.tb03798.x
Edwards, J. A., Chen, C., Kemski, M. M., Hu, J., Mitchell, T. K., and Rappleye, C. A. (2013). Histoplasma yeast and mycelial transcriptomes reveal pathogenic-phase and lineage-specific gene expression profiles. BMC Genomics 14:695. doi: 10.1186/1471-2164-14-695
Fan, W., Kraus, P. R., Boily, M. J., and Heitman, J. (2005). Cryptococcus neoformans gene expression during murine macrophage infection. Eukaryotic Cell 4, 1420–1433. doi: 10.1128/EC.4.8.1420-1433.2005
Garfoot, A. L., Shen, Q., Wüthrich, M., Klein, B. S., and Rappleye, C. A. (2016). The Eng1 β-glucanase enhances Histoplasma virulence by reducing β-Glucan exposure. mBio 7, e01388–e01315. doi: 10.1128/mBio.01388-15
Garfoot, A. L., Zemska, O., and Rappleye, C. A. (2014). Histoplasma capsulatum depends on de novo vitamin biosynthesis for intraphagosomal proliferation. Infect. Immun. 82, 393–404. doi: 10.1128/IAI.00824-13
Hilty, J., George Smulian, A., and Newman, S. L. (2011). Histoplasma capsulatum utilizes siderophores for intracellular iron acquisition in macrophages. Med. Mycol. 49, 633–642. doi: 10.3109/13693786.2011.558930
Holbrook, E. D., Smolnycki, K. A., Youseff, B. H., and Rappleye, C. A. (2013). Redundant catalases detoxify phagocyte reactive oxygen and facilitate Histoplasma capsulatum pathogenesis. Infect. Immun. 81, 2334–2346. doi: 10.1128/IAI.00173-13
Hwang, L., Hocking-Murray, D., Bahrami, A. K., Andersson, M., Rine, J., and Sil, A. (2003). Identifying phase-specific genes in the fungal pathogen Histoplasma capsulatum using a genomic shotgun microarray. Mol. Biol. Cell 14, 2314–2326. doi: 10.1091/mbc.e03-01-0027
Hwang, L. H., Mayfield, J. A., Rine, J., and Sil, A. (2008). Histoplasma requires SID1, a member of an iron-regulated siderophore gene cluster, for host colonization. PLoS Pathog. 4:e1000044. doi: 10.1371/journal.ppat.1000044
Kanetsuna, F., Carbonell, L. M., Azuma, I., and Yamamura, Y. (1972). Biochemical studies on the thermal dimorphism of Paracoccidioides brasiliensis. J. Bacteriol. 110, 208–218. doi: 10.1128/JB.110.1.208-218.1972
Kauffman, C. A. (2009). Histoplasmosis. Clin. Chest Med. 30, 217–225. doi: 10.1016/j.ccm.2009.02.002
Kretschmer, M., Wang, J., and Kronstad, J. W. (2012). Peroxisomal and mitochondrial β-oxidation pathways influence the virulence of the pathogenic fungus Cryptococcus neoformans. Eukaryotic Cell 11, 1042–1054. doi: 10.1128/EC.00128-12
Lacerda Pigosso, L., Baeza, L. C., Vieira Tomazett, M., Batista Rodrigues Faleiro, M., Brianezi Dignani de Moura, V. M., Melo Bailão, A., et al. (2017). Paracoccidioides brasiliensis presents metabolic reprogramming and secretes a serine proteinase during murine infection. Virulence 8, 1417–1434. doi: 10.1080/21505594.2017.1355660
Lorenz, M. C., Bender, J. A., and Fink, G. R. (2004). Transcriptional response of Candida albicans upon internalization by macrophages. Eukaryotic Cell 3, 1076–1087. doi: 10.1128/EC.3.5.1076-1087.2004
Lorenz, M. C., and Fink, G. R. (2001). The glyoxylate cycle is required for fungal virulence. Nature 412, 83–86. doi: 10.1038/35083594
Maresca, B., and Kobayashi, G. S. (1989). Dimorphism in Histoplasma capsulatum: a model for the study of cell differentiation in pathogenic fungi. Microbiol. Rev. 53, 186–209. doi: 10.1128/MMBR.53.2.186-209.1989
Maresca, B., Lambowitz, A. M., Kumar, V. B., Grant, G. A., Kobayashi, G. S., and Medoff, G. (1981). Role of cysteine in regulating morphogenesis and mitochondrial activity in the dimorphic fungus Histoplasma capsulatum. Proc. Natl. Acad. Sci. U.S.A. 78, 4596–4600. doi: 10.1073/pnas.78.7.4596
Medoff, G., Kobayashi, G. S., Painter, A., and Travis, S. (1987). Morphogenesis and pathogenicity of Histoplasma capsulatum. Infect. Immun. 55, 1355–1358. doi: 10.1128/IAI.55.6.1355-1358.1987
Newman, S. L., Bucher, C., Rhodes, J., and Bullock, W. E. (1990). Phagocytosis of Histoplasma capsulatum yeasts and microconidia by human cultured macrophages and alveolar macrophages. Cellular cytoskeleton requirement for attachment and ingestion. J. Clin. Invest. 85, 223–230. doi: 10.1172/JCI114416
Newman, S. L., Gootee, L., Brunner, G., and Deepe, G. S. (1994). Chloroquine induces human macrophage killing of Histoplasma capsulatum by limiting the availability of intracellular iron and is therapeutic in a murine model of histoplasmosis. J. Clin. Invest. 93, 1422–1429. doi: 10.1172/JCI117119
Parente, A. F. A., Bailão, A. M., Borges, C. L., Parente, J. A., Magalhães, A. D., Ricart, C. A. O., et al. (2011). Proteomic analysis reveals that iron availability alters the metabolic status of the pathogenic fungus Paracoccidioides brasiliensis. PLoS ONE 6:e22810. doi: 10.1371/journal.pone.0022810
Parente-Rocha, J. A., Parente, A. F. A., Baeza, L. C., Bonfim, S. M. R. C., Hernandez, O., McEwen, J. G., et al. (2015). Macrophage interaction with Paracoccidioides brasiliensis yeast cells modulates fungal metabolism and generates a response to oxidative stress. PLoS ONE 10:e0137619. doi: 10.1371/journal.pone.0137619
Queiroz-Telles, F., and Escuissato, D. L. (2011). Pulmonary paracoccidioidomycosis. Semin. Respir. Crit. Care Med. 32, 764–774. doi: 10.1055/s-0031-1295724
Rappleye, C. A., Eissenberg, L. G., and Goldman, W. E. (2007). Histoplasma capsulatum alpha-(1,3)-glucan blocks innate immune recognition by the beta-glucan receptor. Proc. Natl. Acad. Sci. U.S.A. 104, 1366–1370. doi: 10.1073/pnas.0609848104
Rappleye, C. A., Engle, J. T., and Goldman, W. E. (2004). RNA interference in Histoplasma capsulatum demonstrates a role for alpha-(1,3)-glucan in virulence. Mol. Microbiol. 53, 153–165. doi: 10.1111/j.1365-2958.2004.04131.x
Rodrigues-Vendramini, F. A. V., Marschalk, C., Toplak, M., Macheroux, P., Bonfim-Mendonça, P., Svidzinski, T. I. E., et al. (2019). Promising new antifungal treatment targeting chorismate synthase from Paracoccidioides brasiliensis. Antimicrob. Agents Chemother. 63, e01097–18. doi: 10.1128/AAC.01097-18
Salvin, S. B. (1949). Cysteine and related compounds in the growth of the yeast like phase of Histoplasma capsulatum. J. Infect. Dis. 84, 275–283. doi: 10.1093/infdis/84.3.275
Salzer, H. J. F., Burchard, G., Cornely, O. A., Lange, C., Rolling, T., Schmiedel, S., et al. (2018). Diagnosis and management of systemic endemic mycoses causing pulmonary disease. Respiration 96, 283–301. doi: 10.1159/000489501
San-Blas, G., San-Blas, F., and Serrano, L. E. (1977). Host-parasite relationships in the yeastlike form of Paracoccidioides brasiliensis strain IVIC Pb9. Infect. Immun. 15, 343–346. doi: 10.1128/IAI.15.2.343-346.1977
Shen, Q., Beucler, M. J., Ray, S. C., and Rappleye, C. A. (2018). Macrophage activation by IFN-γ triggers restriction of phagosomal copper from intracellular pathogens. PLoS Pathog. 14:e1007444. doi: 10.1371/journal.ppat.1007444
Shen, Q., and Rappleye, C. A. (2017). Differentiation of the fungus Histoplasma capsulatum into a pathogen of phagocytes. Curr. Opin. Microbiol. 40, 1–7. doi: 10.1016/j.mib.2017.10.003
Shen, Q., Ray, S. C., Evans, H. M., Deepe, G. S., and Rappleye, C. A. (2020). Metabolism of gluconeogenic substrates by an intracellular fungal pathogen circumvents nutritional limitations within macrophages. mBio 11, e02712–19. doi: 10.1128/mBio.02712-19
Silva-Bailão, M. G., Bailão, E. F. L. C., Lechner, B. E., Gauthier, G. M., Lindner, H., Bailão, A. M., et al. (2014). Hydroxamate production as a high affinity iron acquisition mechanism in Paracoccidioides spp. PLoS ONE 9:e105805. doi: 10.1371/journal.pone.0105805
Soares, D. A., de Andrade, R. V., Silva, S. S., Bocca, A. L., Soares Felipe, S. M., and Petrofeza, S. (2010). Extracellular Paracoccidioides brasiliensis phospholipase B involvement in alveolar macrophage interaction. BMC Microbiol. 10:241. doi: 10.1186/1471-2180-10-241
Souto, J. T., Figueiredo, F., Furlanetto, A., Pfeffer, K., Rossi, M. A., and Silva, J. S. (2000). Interferon-gamma and tumor necrosis factor-alpha determine resistance to Paracoccidioides brasiliensis infection in mice. Am. J. Pathol. 156, 1811–1820. doi: 10.1016/S0002-9440(10)65053-5
Stetler, D. A., and Boguslawski, G. (1979). Cysteine biosynthesis in a fungus, Histoplasma capsulatum. Sabouraudia 17, 23–34. doi: 10.1080/00362177985380041
Subramanian Vignesh, K., Landero Figueroa, J. A., Porollo, A., Caruso, J. A., and Deepe, G. S. (2013). Granulocyte macrophage-colony stimulating factor induced Zn sequestration enhances macrophage superoxide and limits intracellular pathogen survival. Immunity 39, 697–710. doi: 10.1016/j.immuni.2013.09.006
Tavares, A. H. F. P., Silva, S. S., Dantas, A., Campos, E. G., Andrade, R. V., Maranhão, A. Q., et al. (2007). Early transcriptional response of Paracoccidioides brasiliensis upon internalization by murine macrophages. Microbes Infect. 9, 583–590. doi: 10.1016/j.micinf.2007.01.024
Tristão, F. S. M., Rocha, F. A., Carlos, D., Ketelut-Carneiro, N., Souza, C. O. S., Milanezi, C. M., et al. (2017). Th17-inducing cytokines IL-6 and IL-23 are crucial for granuloma formation during experimental paracoccidioidomycosis. Front. Immunol. 8:949. doi: 10.3389/fimmu.2017.00949
Van Dyke, M. C. C., Teixeira, M. M., and Barker, B. M. (2019). Fantastic yeasts and where to find them: the hidden diversity of dimorphic fungal pathogens. Curr. Opin. Microbiol. 52, 55–63. doi: 10.1016/j.mib.2019.05.002
Keywords: Histoplasma, Paracoccidioides, macrophage, phagosome, metabolism, fungal pathogen, gluconeogenesis, nutritional immunity
Citation: Shen Q and Rappleye CA (2020) Living Within the Macrophage: Dimorphic Fungal Pathogen Intracellular Metabolism. Front. Cell. Infect. Microbiol. 10:592259. doi: 10.3389/fcimb.2020.592259
Received: 06 August 2020; Accepted: 15 September 2020;
Published: 16 October 2020.
Edited by:
Angel Gonzalez, University of Antioquia, ColombiaReviewed by:
Flavio Vieira Loures, Federal University of São Paulo, BrazilJulhiany De Fátima Da Silva, Universidade Estadual Paulista, Brazil
Copyright © 2020 Shen and Rappleye. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Chad A. Rappleye, rappleye.1@osu.edu
 Qian Shen
Qian Shen Chad A. Rappleye
Chad A. Rappleye