The Biochemical and Genetic Basis for the Biosynthesis of Bioactive Compounds in Hypericum perforatum L., One of the Largest Medicinal Crops in Europe
Abstract
:1. Introduction
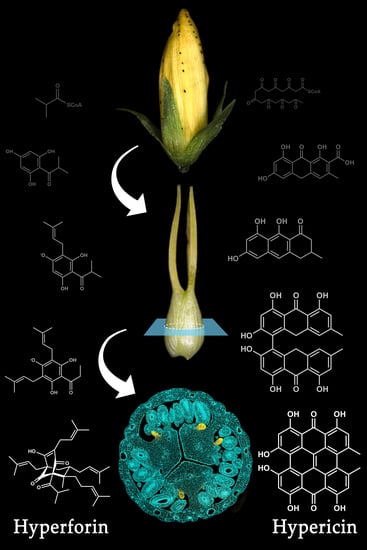
2. The Relevance of Hyperforin, Hypericin and Other Bioactive Compounds from Hypericum perforatum L.
3. Secretory Structures of Hypericum perforatum
4. The Frontier of Dark Gland and Hypericin Biosynthesis Research
5. Hypericin Biosynthesis
6. Hyperforin Biosynthesis
7. Perspectives of Engineering the Biosynthetic Pathways of Relevant Compounds from H. perforatum in Microorganisms and Plants
8. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Crockett, S.L.; Robson, N.K.B. Taxonomy and Chemotaxonomy of the Genus Hypericum. Med. Aromat. Plant Sci. Biotechnol. 2011, 5, 1–13. [Google Scholar] [PubMed]
- Oliveira, A.I.; Pinho, C.; Sarmento, B.; Dias, A.C.P. Neuroprotective activity of hypericum perforatum and its major components. Front. Plant Sci. 2016, 7, 1–15. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Barnes, J.; Anderson, L.A.; Phillipson, J.D. St John’s wort (Hypericum perforatum L.): A Review of its Chemistry, Pharmacology and Clinical properties. J. Pharmacol. Sci. 2001, 583–600. [Google Scholar] [CrossRef]
- Müller, W.E. St. John’s Wort and Its Active Principle in Depression and Anxiety; Springer Science & Business Media: Berlin/Heidelberg, Germany, 2005. [Google Scholar]
- Bhattacharya, S.K.; Chakrabarti, A.; Chatterjee, S.S. Activity profiles of two hyperforin-containing hypericum extracts in behavioral models. Pharmacopsychiatry 1998, 31, 22–29. [Google Scholar] [CrossRef] [PubMed]
- Chatterjee, S.S.; Bhattacharya, S.K.; Wonnemann, M.; Singer, A.; Müller, W.E. Hyperforin as a possible antidepressant component of Hypericum extracts. Life Sci. 1998, 63, 499–510. [Google Scholar] [CrossRef]
- Schellenberg, R.; Sauer, S.; Dimpfel, W. Pharmacodynamic effects of two different hypericum extracts in healthy volunteers measured by quantitative EEG. Pharmacopsychiatry 1998, 31, 44–53. [Google Scholar] [CrossRef]
- Dimpfel, W.; Schober, F.; Mannel, M. Effects of a methanolic extract and a hyperforin-enriched CO2 extract of St. John’s Wort (Hypericum perforatum) on intracerebral field potentials in the freely moving rat (Tele-Stereo-EEG). Pharmacopsychiatry 1998, 31, 30–35. [Google Scholar] [CrossRef]
- Klemow, M.K.; Bartlow, A.; Crawford, J.; Kocher, N.; Shah, J.; Ritsick, M. Herbal Medicine Biomolecular and Clinical Aspects, 2nd ed.; Benzie, I.F.F., Wachtel-Galor, S., Eds.; CRC Press: Boca Raton, FL, USA, 2011; Volume 9, ISBN 9781439807132. [Google Scholar]
- Hofrichter, J.; Krohn, M.; Schumacher, T.; Lange, C.; Feistel, B.; Walbroel, B.; Heinze, H.-J.; Crockett, S.; Sharbel, T.F.; Pahnke, J. Reduced Alzheimer’s disease pathology by St. John’s Wort treatment is independent of hyperforin and facilitated by ABCC1 and microglia activation in mice. Curr. Alzheimer Res. 2013, 10, 1057–1069. [Google Scholar] [CrossRef]
- Garg, A.D.; Krysko, D.V.; Vandenabeele, P.; Agostinis, P. Hypericin-based photodynamic therapy induces surface exposure of damage-associated molecular patterns like HSP70 and calreticulin. Cancer Immunol. Immunother. 2012, 61, 215–221. [Google Scholar] [CrossRef]
- Karppinen, K.; Hokkanen, J.; Mattila, S.; Neubauer, P.; Hohtola, A. Octaketide-producing type III polyketide synthase from Hypericum perforatum is expressed in dark glands accumulating hypericins. FEBS J. 2008, 275, 4329–4342. [Google Scholar] [CrossRef]
- Soták, M.; Czeranková, O.; Klein, D.; Jurčacková, Z.; Li, L.; Čellárová, E. Comparative transcriptome reconstruction of four hypericum species focused on Hypericin biosynthesis. Front. Plant Sci. 2016, 7, 1–14. [Google Scholar] [CrossRef] [Green Version]
- Rizzo, P.; Altschmied, L.; Stark, P.; Rutten, T.; Gündel, A.; Scharfenberg, S.; Franke, K.; Bäumlein, H.; Wessjohann, L.; Koch, M.; et al. Discovery of key regulators of dark gland development and hypericin biosynthesis in St. John’s Wort (Hypericum perforatum). Plant Biotechnol. J. 2019, 17, 2299–2312. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chandrasekera, D.H.; Heinrich, M.; Ashton, D.; Welham, K.J.; Middleton, R. Quantitative analysis of the major constituents of St John’s wort with HPLC-ESI-MS. J. Pharm. Pharmacol. 2005, 57, 1645–1652. [Google Scholar] [CrossRef]
- Scotti, F.; Löbel, K.; Booker, A.; Heinrich, M. St. John’s Wort (Hypericum perforatum) Products – How Variable Is the Primary Material? Front. Plant Sci. 2019, 9, 1–12. [Google Scholar] [CrossRef] [Green Version]
- Nahrstedt, A.; Butterweck, V. Biologically active and other chemical constituents of the herb of Hypericum perforation L. Pharmacopsychiatry 1997, 30, 129–134. [Google Scholar] [CrossRef] [PubMed]
- Laakmann, G.; Schüle, C.; Baghai, T.; Kieser, M. St. John’s Wort in mild to moderate depression: The relevance of hyperforin for the clinical efficacy. Pharmacopsychiatry 1998, 31, 54–59. [Google Scholar] [CrossRef] [PubMed]
- Cervo, L.; Rozio, M.; Ekalle-Soppo, C.B.; Guiso, G.; Morazzoni, P.; Caccia, S. Role of hyperforin in the antidepressant-like activity of Hypericum perforatum extracts. Psychopharmacology 2002, 164, 423–428. [Google Scholar] [CrossRef]
- Obata, H. Analgesic mechanisms of antidepressants for neuropathic pain. Int. J. Mol. Sci. 2017, 18, 2483. [Google Scholar] [CrossRef] [Green Version]
- Müller, W.E.; Singer, A.; Wonnemann, M. Hyperforin—Antidepressant activity by a novel mechanism of action. Pharmacopsychiatry 2001, 34, 98–102. [Google Scholar] [CrossRef]
- Sneddon, J.M. Sodium-dependent accumulation of 5-hydroxytryptamine by rat blood platelets. Br. J. Pharmacol. 1969, 37, 680–688. [Google Scholar] [CrossRef] [Green Version]
- Bogdanski, D.F.; Tissari, A.H.; Brodie, B. Effects of sodium and potassium on kinetics of 5-hydroxytryptamine and norepinephrine transport by rabbit synaptosomes. Biochim. Biophys. Acta 1970, 219, 189–199. [Google Scholar] [CrossRef]
- Shelton, R.C.; Keller, M.B.; Gelenberg, A.; Dunner, D.L.; Hirschfeld, R.; Thase, M.E.; Russel, J.; Lydiard, R.B.; Crits-Cristoph, P.; Gallop, R.; et al. Effectiveness of St. John’s Wort in major depression: A randomized controlled trial. J. Am. Med. Assoc. 2001, 285, 1978–1987. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hypericum Depression Trial Study Group Effects of Hypericum perforatum (St John’s Wort) in Major Depressive Disorder. J. Am. Med. Assoc. 2002, 287, 1807–1814. [CrossRef] [PubMed] [Green Version]
- Linde, K.; Berner, M.M.; Kriston, L. St John’s wort for major depression (Review). Wiley Cochrane Collab. 2008, 4, 1–55. [Google Scholar]
- Schempp, C.M.; Pelz, K.; Wittmer, A.; Schöpf, E.; Simon, J.C. Antibacterial activity of hyperforin from St John’s wort, against multiresistant Staphylococcus aureus and gram-positive bacteria. Lancet 1999, 353, 2129. [Google Scholar] [CrossRef]
- Dudek-Perić, A.M.; Gołąb, J.; Garg, A.D.; Agostinis, P. Melanoma targeting with the loco-regional chemotherapeutic, Melphalan: From cell death to immunotherapeutic efficacy. Oncoimmunology 2015, 4, 5–7. [Google Scholar] [CrossRef] [Green Version]
- Agostinis, P.; Assefa, Z.; Vantieghem, A.; Vandenheede, J.R.; Merlevede, W.; De Witte, P. Apoptotic and anti-apoptotic signaling pathways induced by photodynamic therapy with hypericin. Adv. Enzyme Regul. 2000, 40, 157–182. [Google Scholar] [CrossRef]
- Agostinis, P.; Vantieghem, A.; Merlevede, W.; De Witte, P.A.M. Hypericin in cancer treatment: More light on the way. Int. J. Biochem. Cell Biol. 2002, 34, 221–241. [Google Scholar] [CrossRef]
- Garg, A.D.; Nowis, D.; Golab, J.; Agostinis, P. Photodynamic therapy: Illuminating the road from cell death towards anti-tumour immunity. Apoptosis 2010, 15, 1050–1071. [Google Scholar] [CrossRef]
- Krysko, O.; Løve Aaes, T.; Bachert, C.; Vandenabeele, P.; Krysko, D. V Many faces of DAMPs in cancer therapy. Cell Death Dis. 2013, 4, e631. [Google Scholar] [CrossRef] [Green Version]
- Chen, H.; Muhammad, I.; Zhang, Y.; Ren, Y.; Zhang, R.; Huang, X.; Diao, L.; Liu, H.; Li, X.; Sun, X.; et al. Antiviral activity against infectious bronchitis virus and bioactive components of Hypericum perforatum L. Front. Pharmacol. 2019, 10, 1–22. [Google Scholar] [CrossRef]
- Degar, S.; Prince, A.M.; Pascual, D.; Lavie, G.; Levin, B.; Mazur, Y.; Lavie, D.; Ehrlich, L.S.; Carter, C.; Meruelo, D. Inactivation of the Human Immunodeficiency Virus by Hypericin: Evidence for Photochemical Alterations of p24 and a Block in Uncoating. AIDS Res. Hum. Retroviruses 1992, 8, 1929–1936. [Google Scholar] [CrossRef] [PubMed]
- Hudson, J.B.; Harris, L.; Towers, G.H.N. The importance of light in the anti-HIV effect of hypericin. Antiviral Res. 1993, 20, 173–178. [Google Scholar] [CrossRef]
- Tang, J.; Colacino, J.M.; Larsen, S.H.; Spitzer, W. Virucidal activity of hypericin against enveloped and non-enveloped DNA and RNA viruses. Antiviral Res. 1990, 13, 313–325. [Google Scholar] [CrossRef]
- Prince, A.M.; Pascual, D.; Meruelo, D.; Liebes, L.; Mazur, Y.; Dubovi, E.; Mandel, M.; Lavie, G. Strategies for Evaluation of Enveloped Virus Inactivation in Red Cell Concentrates Using Hypericin. Photochem. Photobiol. 2000, 71, 188–195. [Google Scholar] [CrossRef]
- Shih, C.M.; Wu, C.H.; Wu, W.J.; Hsiao, Y.M.; Ko, J.L. Hypericin inhibits hepatitis C virus replication via deacetylation and down-regulation of heme oxygenase-1. Phytomedicine 2018, 46, 193–198. [Google Scholar] [CrossRef] [PubMed]
- Du, X.; Xiao, R.; Fu, H.; Yuan, Z.; Zhang, W.; Yin, L.; He, C.; Li, C.; Zhou, J.; Liu, G.; et al. Hypericin-loaded graphene oxide protects ducks against a novel duck reovirus. Mater. Sci. Eng. C 2019, 105, 110052. [Google Scholar] [CrossRef] [PubMed]
- Mehjabin, R.; Xiong, L.; Huang, R.; Yang, C.; Chen, G.; He, L.; Liao, L.; Zhu, Z.; Wang, Y. Full-length transcriptome sequencing and the discovery of new transcripts in the unfertilized eggs of Zebrafish (Danio rerio). G3 Genes Genomes Genet. 2019, 9, 1831–1838. [Google Scholar] [CrossRef] [Green Version]
- Lenard, J.; Rabsont, A.; Vanderoef, R. Photodynamic inactivation of infectivity of human immunodeficiency virus and other enveloped viruses using hypericin and rose bengal: Inhibition of fusion and syncytia formation (vesicular stomatitis virus/influenza virus/Sendai virus/hemolysis). Proc. Natl. Acad. Sci. USA 1993, 90, 158–162. [Google Scholar] [CrossRef] [Green Version]
- Sgarbossa, A.; Buselli, D.; Lenci, F. In vitro perturbation of aggregation processes in β-amyloid peptides: A spectroscopic study. FEBS Lett. 2008, 582, 3288–3292. [Google Scholar] [CrossRef] [Green Version]
- Bramanti, E.; Lenci, F.; Sgarbossa, A. Effects of hypericin on the structure and aggregation properties of β-amyloid peptides. Eur. Biophys. J. 2010, 39, 1493–1501. [Google Scholar] [CrossRef] [PubMed]
- Talaga, P. Inhibitors of β-amyloid aggregation: Still an issue of structure and function? Drug Discov. Today 2004, 1, 7–12. [Google Scholar] [CrossRef]
- Chimon, S.; Shaibat, M.A.; Jones, C.R.; Calero, D.C.; Aizezi, B.; Ishii, Y. Evidence of fibril-like β-sheet structures in a neurotoxic amyloid intermediate of Alzheimer’s β-amyloid. Nat. Struct. Mol. Biol. 2007, 14, 1157–1164. [Google Scholar] [CrossRef]
- Zou, Y.; Lu, Y.; Wei, D. Antioxidant activity of a flavonoid-rich extract of Hypericum perforatum L. in vitro. J. Agric. Food Chem. 2004, 52, 5032–5039. [Google Scholar] [CrossRef] [PubMed]
- Silva, B.; Oliveira, P.J.; Dias, A.; Malva, J.O. Quercetin, kaempferol and biapigenin from hypericum perforatum are neuroprotective against excitotoxic insults. Neurotox. Res. 2008, 13, 265–279. [Google Scholar] [CrossRef]
- Soelberg, J.; Jørgensen, L.B.; Jäger, A.K. Hyperforin accumulates in the translucent glands of Hypericum perforatum. Ann. Bot. 2007, 99, 1097–1100. [Google Scholar] [CrossRef] [Green Version]
- Ciccarelli, D.; Andreucci, A.C.; Pagni, A.M. Translucent Glands and Secretory Canals in Hypericum perforatum L. (Hypericaceae): Morphological, Anatomical and Histochemical Studies During the Course of Ontogenesis. Ann. Bot. 2001, 88, 637–644. [Google Scholar] [CrossRef] [Green Version]
- Zobayed, S.M.A.; Afreen, F.; Goto, E.; Kozai, T. Plant-environment interactions: Accumulation of hypericin in dark glands of Hypericum perforatum. Ann. Bot. 2006, 98, 793–804. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Onelli, E.; Rivetta, A.; Giorgi, A.; Bignami, M.; Cocucci, M.; Patrignani, G. Ultrastructural studies on the developing secretory nodules of Hypericum perforatum. Flora 2002, 197, 92–102. [Google Scholar] [CrossRef]
- Soták, M.; Czeranková, O.; Klein, D.; Nigutová, K.; Altschmied, L.; Li, L.; Adarsch, J.; Wurtele, E.S.; Cellarova, E. Differentially Expressed Genes in Hypericin-Containing Hypericum perforatum Leaf Tissues as Revealed by De Novo Assembly of RNA-Seq. Plant Mol. Biol. Rep. 2016, 1027–1041. [Google Scholar] [CrossRef]
- Stracke, R.; Werber, M.; Weisshaar, B. The R2R3-MYBgene family in Arabidopsis thaliana. Cell Signal. Gene Regul. 2001, 447–456. [Google Scholar] [CrossRef]
- Feng, C.; Andreasson, E.; Maslak, A.; Mock, H.P.; Mattsson, O.; Mundy, J. Arabidopsis MYB68 in development and responses to environmental cues. Plant Sci. 2004, 167, 1099–1107. [Google Scholar] [CrossRef]
- Müller, D.; Schmitz, G.; Theres, K. Blind homologous R2R3 Myb genes control the pattern of lateral meristem initiation in Arabidopsis. Plant Cell 2006, 18, 586–597. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fujiwara, S.; Mitsuda, N.; Nakai, Y.; Kigoshi, K.; Suzuki, K.; Ohme-Takagi, M. Chimeric repressor analysis identifies MYB87 as a possible regulator of morphogenesis via cell wall organization and remodeling in Arabidopsis. Biotechnol. Lett. 2014, 36, 1049–1057. [Google Scholar] [CrossRef] [PubMed]
- Liberman, L.M.; Sparks, E.E.; Moreno-risueno, M.A.; Petricka, J.J.; Benfey, P.N. MYB36 regulates the transition from proliferation to differentiation in the Arabidopsis root. Proc. Natl. Acad. Sci. USA 2015, 112, 12099–12104. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ohmori, S.; Kimizu, M.; Sugita, M.; Miyao, A.; Hirochika, H.; Uchida, E.; Nagato, Y.; Yoshida, H. MOSAIC FLORAL ORGANS1, an AGL6-Like MADS Box Gene, Regulates Floral Organ Identity and Meristem Fate in Rice. Plant Cell 2009, 21, 3008–3025. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Viaene, T.; Vekemans, D.; Becker, A.; Melzer, S.; Geuten, K. Expression divergence of the AGL6 MADS domain transcription factor lineage after a core eudicot duplication suggests functional diversification. BMC Plant Biol. 2010, 10, 148. [Google Scholar] [CrossRef] [Green Version]
- Zhao, J. Flavonoid transport mechanisms: How to go, and with whom. Trends Plant Sci. 2015, 20, 576–585. [Google Scholar] [CrossRef]
- Buchner, A. Buchner’s Report. Pharmacie 1830, 217. [Google Scholar]
- Černý, C. Über das Hypericin (Hypericumrot). Zeitschrift für Physiol. Chemie 1911, 73, 371–381. [Google Scholar] [CrossRef] [Green Version]
- Brockmann, H.; Kluge, F.; Muxfeldt, H. Totalsynthese des Hypericins. Chem. Ber. 1957, 37, 2302–2318. [Google Scholar] [CrossRef]
- Brockmann, H.; Pohl, F.; Maier, K.; Haschad, M. Über das Hypericin, den photodynamischen Farbstoff des Johanniskrautes (Hypericum perforatum). Justus Liebigs Annalen der Chemie. Justus Liebigs Ann. Chem. 1942, 553, 1–53. [Google Scholar] [CrossRef]
- Brockmann, H.; Falkenhausen, E.; Neeff, R.; Dorlars, A.; Budde, G. Die Konstitution des Hypericins. Chem. Ber. 1951, 84, 865–887. [Google Scholar] [CrossRef]
- Falk, H. From the Photosensitizer Hypericin to the Photoreceptor Stentorin- The Chemistry of Phenanthroperylene Quinones. Angew. Chemie 1999, 38, 3116–3136. [Google Scholar] [CrossRef]
- Agarwal, P.; Agarwal, P.K. Pathogenesis related-10 proteins are small, structurally similar but with diverse role in stress signaling. Mol. Biol. Rep. 2014, 599–611. [Google Scholar] [CrossRef] [PubMed]
- Bais, H.P.; Vepachedu, R.; Lawrence, C.B.; Stermitz, F.R.; Vivanco, J.M. Molecular and biochemical characterization of an enzyme responsible for the formation of hypericin in St. John’s wort (Hypericum perforatum L.). J. Biol. Chem. 2003, 278, 32413–32422. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Brockmann, H.; Sanne, W. Zur Biosynthese des Hypericins. Naturwissenschaften 1953, 40, 509–510. [Google Scholar] [CrossRef]
- Gill, M.; Gimenez, A.; Mckenzie, R.W. Pigments of Fungi, Part 8. Bianthroquinones from Democybe austroveneta. J. Nat. Prod. 1988, 51, 1251–1256. [Google Scholar] [CrossRef]
- Kosuth, J.; Katkovcinova, Z.; Olexova, P.; Cellarova, E. Expression of the hyp-1 gene in early stages of development of Hypericum perforatum L. Plant Cell Rep. 2007, 211–217. [Google Scholar] [CrossRef]
- Michalska, K.; Fernandes, H.; Sikorski, M.; Jaskolski, M. Crystal structure of Hyp-1, a St. John’s wort protein implicated in the biosynthesis of hypericin. J. Struct. Biol. 2010, 169, 161–171. [Google Scholar] [CrossRef]
- Kosuth, J.; Smelcerovic, A.; Borsch, T.; Zuehlke, S.; Karppinen, K.; Spiteller, M.; Hohtola, A.; Cellárová, E. The hyp-1 gene is not a limiting factor for hypericin biosynthesis in the genus Hypericum. Funct. Plant Biol. 2011, 35–43. [Google Scholar] [CrossRef]
- Kosuth, J.; Hrehorova, D.; Jaskolski, M.; Cellarova, E. Stress-induced expression and structure of the putative gene hyp-1 for hypericin biosynthesis. Plant Cell Tissue Org. Cult. 2013, 114, 207–216. [Google Scholar] [CrossRef]
- Karppinen, K.; Derzsó, E.; Jaakola, L.; Hohtola, A. Molecular Cloning and Expression Analysis of hyp-1 Type PR-10 Family Genes in Hypericum perforatum. Front. Plant Sci. 2016, 7, 1–12. [Google Scholar] [CrossRef] [Green Version]
- Sliwiak, J.; Dauter, Z.; Jaskolski, M. Crystal Structure of Hyp-1, a Hypericum perforatum PR-10 Protein, in Complex with Melatonin. Front. Plant Sci. 2016, 7, 1–10. [Google Scholar] [CrossRef] [Green Version]
- Mogensen, J.E.; Wimmer, R.; Larsen, J.N.; Spangfort, M.D.; Otzen, D.E. The Major Birch Allergen, Bet v 1, Shows Affinity for a Broad Spectrum of Physiological Ligands. J. Biol. Chem. 2002, 277, 23684–23692. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lee, E.-J.; Facchini, P. Norcoclaurine Synthase Is a Member of the Pathogenesis-Related 10/Bet v1 Protein Family. Plant Cell 2010, 22, 3489–3503. [Google Scholar] [CrossRef] [Green Version]
- Karppinen, K.; Hohtola, A. Molecular cloning and tissue-specific expression of two cDNAs encoding polyketide synthases from Hypericum perforatum. J. Plant Physiol. 2008, 165, 1079–1086. [Google Scholar] [CrossRef]
- Kusari, S.; Sezgin, S.; Nigutova, K.; Cellarova, E.; Spiteller, M. Spatial chemo-profiling of hypericin and related phytochemicals in Hypericum species using MALDI-HRMS imaging. Anal. Bioanal. Chem. 2015, 407, 4779–4791. [Google Scholar] [CrossRef] [PubMed]
- Kucharíková, A.; Kimáková, K.; Janfelt, C.; Čellárová, E. Interspecific variation in localization of hypericins and phloroglucinols in the genus Hypericum as revealed by desorption electrospray ionization mass spectrometry imaging. Physiol. Plant. 2016, 157, 2–12. [Google Scholar] [CrossRef]
- Abe, I.; Oguro, S.; Utsumi, Y.; Sano, Y.; Noguchi, H. Engineered Biosynthesis of Plant Polyketides: Chain Length Control in an Octaketide-Producing Plant Type III Polyketide Synthase. J. Am. Chem. Soc. 2005, 127, 12709–12716. [Google Scholar] [CrossRef] [PubMed]
- Mizuuchi, Y.; Shi, S.; Wanibuchi, K.; Kojima, A.; Morita, H.; Noguchi, H.; Abe, I. Novel type III polyketide synthases from Aloe arborescens. FEBS J. 2009, 276, 2391–2401. [Google Scholar] [CrossRef]
- Gagne, S.J.; Stout, J.M.; Liu, E.; Boubakir, Z.; Clark, S.M.; Page, J.E. Identification of olivetolic acid cyclase from Cannabis sativa reveals a unique catalytic route to plant polyketides. Proc. Natl. Acad. Sci. USA 2012, 109, 12811–12816. [Google Scholar] [CrossRef] [Green Version]
- Kimáková, K.; Kimáková, A.; Idkowiak, J.; Stobiecki, M.; Rodziewicz, P. Phenotyping the genus Hypericum by secondary metabolite profiling: Emodin vs. skyrin, two possible key intermediates in hypericin biosynthesis. Anal. Bioanal. Chem. 2018, 410, 7689–7699. [Google Scholar] [CrossRef] [Green Version]
- Bystrov, N.S.; Chernov, B.K.; Dobrynin, V.N.; Kolosov, M.N. The structure of hyperforin. Tetrahedron Lett. 1975, 16, 2791–2794. [Google Scholar] [CrossRef]
- Brondz, I.; Greibrokk, T.; Groth, P.A.; Aasen, A.J. The relative stereochemistry of hyperforin—An antibiotic from L. Tetrahedron Lett. 1982, 23, 1299–1300. [Google Scholar] [CrossRef]
- Adam, P.; Arigoni, D.; Bacher, A.; Eisenreich, W. Biosynthesis of hyperforin in Hypericum perforatum. J. Med. Chem. 2002, 45, 4786–4793. [Google Scholar] [CrossRef] [PubMed]
- Maisenbacher, P.; Kovar, K.-A. Analysis and Stability of Hyperici Oleum. Planta Med. 1992, 58, 351–354. [Google Scholar] [CrossRef]
- Erdelmeier, C. Hyperforin, Possibly the Major Non-Nitrogenous Secondary Metabolite of Hypericum perforatum L. Pharmacopsychiatry 1998, 31, 2–6. [Google Scholar] [CrossRef] [PubMed]
- Verotta, L.; Appendino, G.; Jakupovic, J.; Bombardelli, E. Hyperforin Analogues from St. John’s Wort (Hypericum perforatum). J. Nat. Prod. 2000, 63, 412–415. [Google Scholar] [CrossRef] [PubMed]
- De Shan, M.; Hu, L.H.; Chen, Z.L. Three New Hyperforin Analogues from Hypericum perforatum. J. Nat. Prod. 2001, 64, 127–130. [Google Scholar] [CrossRef]
- Liu, F.; Pan, C.; Drumm, P.; Ang, C.Y.W. Liquid chromatography–mass spectrometry studies of St. John’s wort methanol extraction: Active constituents and their transformation. J. Pharm. Biomed. Anal. 2005, 37, 303–312. [Google Scholar] [CrossRef] [PubMed]
- Brahmachari, G. Biosynthetic and Total Synthetic Approaches for (+)-Hyperforin. In Discovery and Development of Neuroprotective Agents from Natural Products; Elsevier: Amsterdam, The Netherlands, 2018; pp. 435–456. ISBN 9780128097694. [Google Scholar]
- Piovan, A.; Filippini, R.; Caniato, R.; Borsarini, A.; Maleci, L.B.; Cappelletti, E.M. Detection of hypericins in the ‘red glands’ of Hypericum elodes by ESI-MS/MS. Phytochemistry 2004, 65, 411–414. [Google Scholar] [CrossRef] [PubMed]
- Umek, A.; Kreft, S.; Kartnig, T.; Heydel, B. Quantitative Phytochemical Analyses of Six Hypericum Species Growing in Slovenia. Planta Med. 1999, 65, 388–390. [Google Scholar] [CrossRef]
- Tekel’ová, D.; Repčák, M.; Zemková, E.; Tóth, J. Quantitative Changes of Dianthrones, Hyperforin and Flavonoids Content in the Flower Ontogenesis of Hypericum perforatum. Planta Med. 2000, 66, 778–780. [Google Scholar] [CrossRef]
- Maisenbacher, P.; Kovar, K.-A. Adhyperforin: A Homologue of Hyperforin from Hypericum perforatum. Planta Med. 1992, 58, 291–293. [Google Scholar] [CrossRef] [PubMed]
- Grigson, G. The Englishman’s Flora. In The Englishman’s Flora; Harper-Collins: New York, NY, USA, 1958; pp. 83–89. ISBN 0246108207. [Google Scholar]
- Gurevich, A.I.; Dobrynin, V.N.; Kolosov, M.N.; Popravko, S.A.; Riabova, I.D. Antibiotic hyperforin from Hypericum perforatum L. Antibiotiki 1971, 16, 510–513. [Google Scholar] [PubMed]
- Brondz, I.; Greibrokk, T.; Groth, P.; Aasen, A.J.; Seip, R.; Brunvoll, J. The Absolute Configuration of Hyperforin, an Antibiotic from Hypericum perforatum L., Based on the Crystal Structure Determination of its p-Bromobenzoate Ester. Acta Chem. Scand. 1983, 37a, 263–265. [Google Scholar] [CrossRef] [Green Version]
- Zhou, W.; Zhuang, Y.; Bai, Y.; Bi, H.; Liu, T.; Ma, Y. Biosynthesis of phlorisovalerophenone and 4-hydroxy-6-isobutyl-2-pyrone in Escherichia coli from glucose. Microb. Cell Fact. 2016, 15, 149. [Google Scholar] [CrossRef] [Green Version]
- Karppinen, K.; Hokkanen, J.; Tolonen, A. Biosynthesis of hyperforin and adhyperforin from amino acid precursors in shoot cultures of Hypericum perforatum. Phytochemistry 2007, 68, 1038–1045. [Google Scholar] [CrossRef]
- Smetanska, I. Production of secondary metabolites using plant cell cultures. In Food Biotechnology; Stahl, U., Donalies, U.E.B., Nevoigt, E., Eds.; Springer: Berlin/Heidelberg, Germany, 2007; Volume 132, pp. 188–224. ISBN 9783540705352. [Google Scholar]
- Lichtenthaler, H. Sterols and Isoprenoids. Biochem. Soc. Trans. 2000, 28, 785–789. [Google Scholar] [CrossRef]
- Paniego, N.B.; Zuurbier, K.W.M.; Fung, S.Y.; Van Der Heijden, R.; Scheffer, J.J.C.; Verpoorte, R. Phlorisovalerophenone synthase, a novel polyketide synthase from hop (Humulus lupulus L.) cones. Eur. J. Biochem. 1999, 262, 612–616. [Google Scholar] [CrossRef] [PubMed]
- Klingauf, P.; Beuerle, T.; Mellenthin, A.; El-moghazy, S.A.M. Biosynthesis of the hyperforin skeleton in Hypericum calycinum cell cultures. Phytochemistry 2005, 66, 139–145. [Google Scholar] [CrossRef]
- Hillwig, M.L. Understanding the Biological Activity of Hypericum Species through Metabolomics Studies. Ph.D. Thesis, Iowa State University, Ames, IA, USA, 2008. [Google Scholar]
- Jez, J.M.; Bowman, M.E.; Noel, J.P. Expanding the biosynthetic repertoire of plant type III polyketide synthases by altering starter molecule specificity. Proc. Natl. Acad. Sci. USA 2002, 99, 5319–5324. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Abe, I.; Morita, H. Structure and function of the chalcone synthase superfamily of plant type III polyketide synthases. Nat. Prod. Rep. 2010, 27, 809–838. [Google Scholar] [CrossRef]
- Okada, Y.; Yamazaki, Y.; Suh, D.Y.; Sankawa, U.; Ito, K. Bifunctional activities of Valerophenone synthase in hop (Humulus lupulus L.). J. Am. Soc. Brew. Chem. 2001, 59, 163–166. [Google Scholar] [CrossRef]
- Clark, S.M.; Vaitheeswaran, V.; Ambrose, S.J.; Purves, R.W.; Page, J.E. Transcriptome analysis of bitter acid biosynthesis and precursor pathways in hop (Humulus lupulus). BMC Plant Biol. 2013, 13. [Google Scholar] [CrossRef] [Green Version]
- Boubakir, Z.; Beuerle, T.; Liu, B.; Beerhues, L. The first prenylation step in hyperforin biosynthesis. Phytochemistry 2005, 66, 51–57. [Google Scholar] [CrossRef]
- Zuurbier, K.W.M.; Fung, S.Y.; Scheffer, J.J.C.; Verpoorte, R. In-vitro prenylation of aromatic intermediates in the biosynthesis of bitter acids in Humulus lupulus. Phytochemistry 1998, 49, 2315–2322. [Google Scholar] [CrossRef]
- Taura, F.; Tanaka, S.; Taguchi, C.; Fukamizu, T.; Tanaka, H.; Shoyama, Y.; Morimoto, S. Characterization of olivetol synthase, a polyketide synthase putatively involved in cannabinoid biosynthetic pathway. FEBS Lett. 2009, 583, 2061–2066. [Google Scholar] [CrossRef] [Green Version]
- Beerhues, L. Hyperforin. Phytochemistry 2006, 67, 2201–2207. [Google Scholar] [CrossRef]
- Kusari, S.; Lamshöft, M.; Zühlke, S.; Spiteller, M. An Endophytic Fungus from Hypericum perforatum that Produces Hypericin. J. Nat. Prod. 2008, 71, 159–162. [Google Scholar] [CrossRef] [PubMed]
- Zubek, S.; Mielcarek, S.; Turnau, K. Hypericin and pseudohypericin concentrations of a valuable medicinal plant Hypericum perforatum L. are enhanced by arbuscular mycorrhizal fungi. Mycorrhiza 2012, 149–156. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lazzara, S.; Militello, M.; Carrubba, A.; Napoli, E.; Saia, S. Arbuscular mycorrhizal fungi altered the hypericin, pseudohypericin, and hyperforin content in flowers of Hypericum perforatum grown under contrasting P availability in a highly organic substrate. Mycorrhiza 2017, 27, 345–354. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cirak, C.; Aksoy, H.M.; Ayan, A.K.; Saglam, B.; Kevseroglu, K. Enhanced Hypericin Production in Hypericum perforatum and Hypericum pruinatum in Response to Inoculation with Two Fungal Pathogens. Plant Prot. Sci. 2004, 41, 109–114. [Google Scholar] [CrossRef] [Green Version]
- Kraus, G.A.; Nguyen, T.H.; Jeon, I. Synthesis of the core bicyclic system of hyperforin and nemorosone. Tetrahedron Lett. 2003, 44, 659–661. [Google Scholar] [CrossRef]
- Mehta, G.; Bera, M.K. Synthetic studies towards the phloroglucin natural product hyperforin: Construction of the fully prenylated bicyclic core. Tetrahedron Lett. 2009, 50, 3519–3522. [Google Scholar] [CrossRef]
- Li, Y.; Luxenburger, E.; Müller, R. An Alternative Isovaleryl CoA Biosynthetic Pathway Involving a Previously Unknown 3-Methylglutaconyl CoA Decarboxylase. Angew. Chemie Int. Ed. 2013, 52, 1304–1308. [Google Scholar] [CrossRef]
- Hou, W.; Shakya, P.; Franklin, G. A perspective on hypericum perforatum genetic transformation. Front. Plant Sci. 2016, 7, 1–12. [Google Scholar] [CrossRef] [Green Version]
- Khan, S.A.; Verma, P.; Arbat, A.; Gaikwad, S.; Parasharami, V.A. Development of enhanced hypericin yielding transgenic plants and somaclones: High throughput direct organogenesis from leaf and callus explants of Hypericum perforatum. Ind. Crops Prod. 2018, 111, 544–554. [Google Scholar] [CrossRef]







Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Rizzo, P.; Altschmied, L.; Ravindran, B.M.; Rutten, T.; D’Auria, J.C. The Biochemical and Genetic Basis for the Biosynthesis of Bioactive Compounds in Hypericum perforatum L., One of the Largest Medicinal Crops in Europe. Genes 2020, 11, 1210. https://doi.org/10.3390/genes11101210
Rizzo P, Altschmied L, Ravindran BM, Rutten T, D’Auria JC. The Biochemical and Genetic Basis for the Biosynthesis of Bioactive Compounds in Hypericum perforatum L., One of the Largest Medicinal Crops in Europe. Genes. 2020; 11(10):1210. https://doi.org/10.3390/genes11101210
Chicago/Turabian StyleRizzo, Paride, Lothar Altschmied, Beena M. Ravindran, Twan Rutten, and John C. D’Auria. 2020. "The Biochemical and Genetic Basis for the Biosynthesis of Bioactive Compounds in Hypericum perforatum L., One of the Largest Medicinal Crops in Europe" Genes 11, no. 10: 1210. https://doi.org/10.3390/genes11101210