Abstract
The parameterisation process of a previously developed modelling structure of the grapevine-downy mildew pathosystem is described. The model incorporates primary and secondary infections, host crop growth and development, along with a linkage between disease on foliage and disease on clusters. This process-based model was developed with a main objective of understanding the behaviour of the pathosystem under different, variable, environmental conditions, or under climate change. Six scenarios of disease conduciveness were developed in order to capture the range of environmental conditions under which potential downy mildew of grapevine epidemics can develop. These climate scenarios were based on moisture and temperature factors. The scenarios were translated into vectors of parameters for primary and secondary infections in the model. Model testing was performed in three steps: (i) an analysis of potential epidemics was conducted from the literature on grapevine downy mildew in order to delineate the behaviour of the pathosystem under different scenarios; (ii) a simulation experiment was conducted to investigate the response of the model to different patterns of environmental conditions, corresponding to six scenarios of disease conduciveness; and (iii) expected and simulated epidemics under these scenarios were compared. In scenarios, the model mobilised existing quantitative information on downy mildew of grapevine and generated outputs that are congruent with expected patterns of potential epidemic. This study indicates that the model is a reliable tool for simulating accurate and robust potential epidemics of downy mildew of grapevine in a scenario analysis. This can have many applications, such as the understanding of the behaviour of the pathosystem under climate change or when partial host resistance is involved.
Similar content being viewed by others
Introduction
Grapevine downy mildew (DM; caused by Plasmopara viticola) is a major disease of Vitis vinifera, causing qualitative and quantitative losses, especially in temperate climates (e.g., Gessler et al. 2011). A generic process-based model of the grapevine downy mildew epidemics, which incorporates the main phases of the disease, has been previously developed (Bove 2018; Bove et al. 2020). This model incorporates primary and secondary infections, host crop growth and development, and, departing from other existing models on DM, also includes a linkage between disease on foliage and disease on clusters (Savary et al. 2009), accounting for the dual nature (i.e., foliage and clusters) of epidemics of downy mildew of grapevine (Bove et al. 2020).
The model is based on the epidemiological concepts of infection, latency and infectiousness (Vanderplank 1963). Epidemics start on the foliage when the first primary infection occurs in the system at a given onset date, on a disease-free and susceptible site. After a latency period of LP days, infected sites start to produce spores and continue to do so during an infectious period of IP days. During the infectious period, the infectious site constantly produces Rc effective spores per day, where Rc corresponds to the basic infection rate corrected for removals (i.e. tissue removed from the infectious process; Vanderplank 1963). Each effective propagule generates a new infection in turn, which, after LP days, produces Rc new effective propagules per day over IP days, and so on. The model therefore leads to an increase of the total number of downy mildew infections on the foliage. The model also represents disease transmission from the foliage to the clusters. Disease dynamics of foliage and clusters is governed by system (foliage, clusters) size, host crop growth, physiological and disease-induced senescence, susceptibility of plant organs, age of organs, and environmental factors (Bove 2018; Bove et al. 2020). The model is intended as a generic model with a main objective of understanding the behaviour of the pathosystem under different, variable, environmental conditions, or under climate change.
The objectives of the present paper are: i) to describe the parameterisation process of the simulation model, ii) to develop scenarios of disease conduciveness based on weather factors, which capture the range of environmental conditions under which grapevine downy mildew epidemics develop; iii) to perform a simulation experiment to assess how the model reacts in different scenarios of disease conduciveness, and iv) to compare these reactions (i.e., model outputs) to published data on epidemics of downy mildew (i.e. Gobbin et al. 2005; Rossi and Caffi 2012; Rumbou and Gessler 2004). The simulation experiment (Teng et al. 1977), whereby simulations are run according to a formal experimental design, was used to investigate the response of the model to different patterns of environmental conditions, corresponding to six scenarios of disease conduciveness, which include weather factors and different levels of primary inoculum.
Materials and methods
Parameters and hypotheses
Numerical values of parameters and coefficients involved in the model (Bove 2018; Bove et al. 2020) were derived from the available literature. We describe below details on how the literature on grapevine downy mildew was mobilised to parameterise the model for a series of processes: (i) crop growth and development; (ii) primary infections, (iii) secondary infections, and iv) infections on clusters. Acronyms, values, dimensions, and references for parameters are provided in Table 1. Parameter values can be easily modified if additional or new knowledge is made available, rendering the model flexible for future use.
Crop growth and development.
Maximum number of sites of the foliage
The considered system is a single grapevine plant surrounded by similar grapevine plants in term of size, physiology and development, and disease (Bove et al. 2020). The simulation time is 200 days (covering the growing season) with a time step of 1 day (Bove et al. 2020). The dynamics of foliage growth and disease on foliage is represented by the running number of foliage sites. The maximum number of sites, Smax, was calculated as the ratio of the maximum leaf area (LA) of a single grapevine plant (i.e., the size of the considered system) over the area of an individual DM lesion (LS): Smax = LA/LS. We used 3 m2 for LA (Bernizzoni et al. 2009) and 314.16 mm2 for LS (Galet 1977), and thus estimated Smax as 10,000 sites.
Relative rate of growth
Foliage growth is assumed to be logistic, with a relative rate of growth RRG (number of new sites produced per site and per unit time, dimension: [N.N−1.T−1]) of 0.1 (Savary and Willocquet 2014).
Physiological senescence
of leaf tissue is assumed to be a negative exponential process and is represented in the model by a rate of loss of healthy sites RSEN, with dimension [Nsite.T−1], that is to say, by the number of new (physiologically) senesced sites (SEN) formed per time step: RSEN(t) = dx(t)/dt, where x represents the number of sites. RSEN is computed using a relative rate of physiological senescence (RRS, dimension: [N.N−1.T−1]), that is to say, considering the number of new (physiologically) senesced sites formed per healthy site per time step: RRS = dx/dt × 1/x. The number of senesced sites at time t may thus be written as: x(t) = exp(RRS × t). The measured RRS (represented as RRSmax in the model) is 0.01 and was calculated from published curves of vegetative assessment in untreated vineyards (Wermelinger and Koblet 1990), following a numerical integration approach.
Disease-induced senescence
is represented in the model by a rate of disease-induced senescence RSENR, with dimension [N.T−1], that is, by the number of new (disease-induced) senesced sites (SENR) formed per time step. RSENR is computed using a relative rate of disease-induced senescence RRDS, with dimension [N.N−1.T−1], that is to say, considering the number of new (diseased-induced) senesced sites formed per removed site (R) per time step: RRDS = dx/dt × 1/x. A comparison between RRS measured from published curves of vegetative assessment in an untreated vineyard in a season (Jermini et al. 2010a) and RRDS measured from DM disease progress curves in the same vineyard and in the same season (Jermini et al. 2010b) led to estimate the value of RRDS as twice that RRS, i.e. is equal to 0.02.
Development stage
is represented in the model by DVS, which is a dimensionless coefficient. DVS can take three values, 0, 1, and 2, where 0 indicates development stages from bud break to flowering, 1 from flowering to veraison, and 2 from veraison to leaf-fall. DVS increases according to the sum of daily temperatures above the threshold for development, Tbase (Willocquet et al. 2008). The value of Tbase was set to 10 °C (Gutierrez et al. 1985; Wermelinger and Koblet 1990; Winkler et al. 1974). DVS is 0 for a temperature sum (SumT) < 300 °C, that is, from the beginning of the simulation (day of simulation, DOS 0, day of year, DOY 90, April 1st) till DOS 59 (DOY 148, May 29th). DVS is 1 for 300 °C < SumT <700 °C, from DOS 60 (DOY 149, May 30th) to DOS 139 (DOY 228, August 17th). DVS is 2 for SumT >700 °C, from DOS 140 (DOY 229, August 18th) to the end of simulation, DOS 200 (DOY 289, October 17th). The DOY values at which bud break, flowering, veraison, and leaf-fall are assumed to occur in the simulation are consistent with reference to Northern Italy (Cola et al. 2014; Gatti et al. 2016; Poni et al. 2006; Rossi et al. 2008).
Primary infections
Onset date
The day of the first seasonal primary infection is represented in the model by the onset date OD, with dimension [T]. The default value (i.e. numerical value of parameters and coefficients used for the numerical evaluation of the model described in Bove et al. 2020) of OD in the model is 30 DOS, which corresponds to DOY 120 (May 1st). This parameter is consistent with the first seasonal disease symptoms appearance in Northern Italy, which is approximately between the beginning of May and the end of June (Rossi et al. 2008).
Duration of mobilization of the primary inoculum
The variable PD [T] represents the number of days during which primary infections contribute to the epidemic. The default value of PD was set to 60 days from disease onset (DOS 30) (Gobbin et al. 2005; Rossi et al. 2009, Fig. 3b; Rumbou and Gessler 2004, 2006), assuming that primary infections do not occur after DOS 90.
Inflow of primary infections
The number of primary infections per time step is represented in the model by P, with dimension [N.T−1]. P represents the rate of primary infection (RPI) between the onset date (OD) and the end of mobilisation of primary inoculum (OD + PD). The number of primary infections per grapevine plant per day was retrieved from published data (Gobbin et al. 2005; Rumbou and Gessler 2004), from the estimation of the number of single P. viticola genotypes (i.e. lesions from primary infections) in several untreated vineyards over different locations. For example, if 184 single P. viticola genotypes were found on 36 vines, during a sampling period of 22 days (first sampling day DOY 155; last sampling day DOY 177), the number of single genotypes per plant was 184/36 = 5.11. Thus, a number of lesion per day was estimated as 5.11/22 = 0.23 (Gobbin et al. 2005). The default value of P was set to 0.2 (Table 1).
Secondary infections on foliage
Optimum relative rate of secondary infections
The optimum value of the basic infection rate corrected for the removals (Rc, Vanderplank 1963) is represented by RcOPT, with dimension [Nles.Nles−1.T−1]. RcOPT is the number of daughter lesions generated per mother lesion per unit time under optimum environmental conditions on susceptible varieties (Savary et al. 2015). We used the approach of Sun and Zeng (1994) to estimate Rc from disease progress curves:
where p is the latency period, i is the infectious period and r is the apparent infection rate (Vanderplank 1963, 1975).
The apparent rate of disease increase can be calculated as follows:
where x1 and x2 are disease fractions at two successive date t1 and t2, respectively, in the early stage of the epidemic under optimum conditions for the disease. Equation (2) was used to calculate r from published disease progress curves (Carisse 2016; Dagostin et al. 2011; Gessler and Blaise 1992; Jermini et al. 2000) in susceptible and unprotected vineyards and under optimum conditions for disease development. The first two non-zero values were retrieved from these disease progress curves. The optimum value of Rc, i.e. RcOpt, was then estimated according to eq. (1), using p = 5 and i = 20 days. For example, if the first two severity values (range: 0–1) of a disease progress curve are x1 = 0.15 at t1 = 231 DOY and x2 = 0.42 at t2 = 239 DOY, respectively, r is 0.12 day−1 and RcOPT is 0.26 day−1 (Jermini et al. 2000). The default value of RcOpt was set to 0.3 (Table 1).
Modifiers of the relative rate of secondary infections
Modifiers (Loomis and Adams 1983) are used in the model in order to account for major environmental factors that are driving epidemics (Bove 2018; Bove et al. 2020). These factors include the daily temperature variation, the daily moisture (caused by rain or dew), age (and variation in susceptibility) and variety (and varietal susceptibility), and are incorporated in the model so that they modify the optimum value of Rc, RcOPT. These modifiers for temperature, moisture, age, and variety, are respectively represented by the dimensionless parameters RcT, RcW, RcA and RcV, respectively, which may vary between 0 and 1 (Bove 2018; Bove et al. 2020). Rc can then be written as:
Since Rc = N × E (Zadoks and Schein 1979, Eq. 6.25), where N is the relative rate of propagule (spore) production, expressed as propagules (spores) per lesion per day, and E is the effectiveness of each dispersal unit, we can write:
In Eq. (3), the four modifiers RcT, RcW, RcA and RcV account for the effects of temperature, moisture, age and variety on N and E, respectively. The parameterisation of the four modifiers was performed considering each element of Eq. (3) from the available literature and is described as follows.
Modifier of RcOPT for age
The modifier RcA accounts for the susceptibility of healthy sites, which depends on the age of the leaves (ontogenic resistance) and on plant development stage. RcA was parameterised from published data reporting the effects of leaf age on susceptibility to downy mildew, for 20 leaf positions on shoots (Reuveni 1998). We assumed that all leaves on a shoot have the same number of sites. We then considered the relationship between the development stage (DVS) and the number of sites (TOTSITES) in the model at each DVS scale, that is: DVS 0: 100 < TOTSITES <7600; DVS 1: 7800 < TOTSITES <10,000; DVS 2: TOTSITES = 10,000. The median value of TOTSITES at each DVS was calculated and the corresponding number of leaves was estimated: DVS 2: 20 × (average TOTSITES/Smax) = 20 leaves; DVS 1: 20 × 8900/10000 = 17.8 leaves; DVS 0: 20 × 3850/10000 = 7.7 leaves. The average severity values for each DVS was retrieved from the Fig. 3 of Reuveni 1998 and was used to estimate RcA, according to infection efficiency (IE): when DVS = 0 the average severity is 0.81 and RcA is 1; when DVS = 1, the average severity is 0.45 and RcA = (1/0.81) × 0.45 = 0.56; when DVS = 2, the average severity is 0.33 and RcA = (1/81) × 0.33 = 0.39. A linear relationship between RcA and severity in the work reported by Reuveni (1998) was assumed.
Modifiers of RcOPT for temperature
Under optimum temperature conditions (15–20 °C), the reported value for IE is 0.07, whereas under less favourable conditions of T (10 °C), IE is 0.06 (Lalancette 1988). Thus, we set RcT to 1 in favourable temperature conditions, and to 0.8 in less favourable temperature conditions (Table 1).
Modifiers of RcOPT for moisture
Under optimum moisture conditions, RcW is 1, whereas, under less favourable conditions of rain and wetness duration, RcW is set to 0.6, accounting for the effect of moisture on IE and N (Lalancette 1987) (Table 1).
Modifier of RcOPT for variety
The effect of susceptibility (or resistance) of variety on infection is accounted by RcV, which is the product of the relative resistance for infection efficiency (RRIE) and the relative resistance for sporulation (RRSP; Savary and Willocquet 2014):
RcV = RRIE × RRSP.
In a susceptible variety, RcV = 1 (Table 1), that is to say, when the variety is associated with the highest level of infection efficiency and sporulation intensity: reduction in RRIE and/or in RRSP in a partially resistant variety leads to a smaller RcV value.
Latency period
The duration of the latency period is represented by LP, with dimension [T]. The default value of LP was set to 6 (Goidanich 1959; Müller and Sleumer 1934).
Infectious period
The duration of the infectious period is represented in the model by the coefficient IP, with dimension [T]. The default value for IP was set to 15 (Caffi et al. 2013; Kennelly et al. 2005).
Disease on clusters
Leaf-cluster disease transmission coefficient
Disease transmission between leaves and clusters is represented by the coefficient TC. Under the hypothesis that cluster infection results from foliage infection, TC reflects the link between disease on the foliage and disease on clusters, expressed as an increase of disease severity on clusters per unit disease severity on leaves per day, according to the equation (Savary et al. 2009, Eq. 2):
where xc is disease severity on clusters; t is time (number of days); xL is the current disease severity on leaves; (1 − xc) represents the proportion of healthy clusters still available for new infections with dimension [1]; and tc is the disease transmission coefficient between leaves and clusters as defined by Savary et al. (2009) and used in Bove et al. (2020). According to the above equation (Savary et al. 2009), the dimension of tc is [Ndc].[Nds−1].[T−1], where Ndc is the number of diseased clusters, Nds is the number of diseased sites on leaves, and T is the unit time (time step = 1 day). The hypothesis underlying the above equation is that disease increase on clusters is only due to infections from secondary inoculum which originates from leaves. According to the estimates of tc reported by Savary et al. (2009) in untreated plots, we set for the parameter TC a value of 0.01 (Table 1).
Coefficient for cluster susceptibility
The development-dependent variation in cluster susceptibility is represented by the dimensionless coefficient CCS. CCS varies between 0 and 1, where 1 corresponds to full susceptibility (i.e. CCSmax). The maximum cluster susceptibility is expressed between flowering and veraison (DVS =1). Cluster susceptibility to downy mildew is considered negligible before flowering (DVS = 0) and after veraison (DVS = 2) (Table 1; Galet 1977; Kennelly et al. 2005).
Scenarios of disease conduciveness
Six scenarios of disease conduciveness were developed in order to assess the effect of the physical environment on downy mildew epidemics, considering moisture and temperature as main weather factors (Gessler et al. 2011). Each of the six weather-related scenarios was defined by different levels of moisture and temperature, and affects the values of seven epidemiological parameters, three of which affect the rate of primary infection, while four affect the rate of secondary infections. The values for the seven parameters in each scenario are given in Table 2.
Weather factors
The considered weather factors were moisture and temperature, at four and two levels, respectively with six combinations (Table 2). The factor accounting for moisture refers to leaf wetness, rainfall, and dew. The four levels of moisture were termed as: “wet”, “partly wet”, “dry” and “very dry”. The two levels of temperature were termed as “warm” and “cold”. Under field conditions, the “wet” level is assumed to coincide with the occurrence of moisture (rain or dew) on the canopy, and corresponds to daily rainfall >5 mm; minimum relative humidity (RHmin) > 70% and maximum relative humidity (RHmax) ≥ 95%. Very dry conditions are defined by RHmin and RHmax below 70% and 95%, respectively, and no rain. The intermediate condition “partly wet” is defined by rainfall between 0 and 5 mm, RHmin ≥70% and RHmax ≤95%. The “dry” level is described by absence of rain, RHmin ≥70% and RHmax ≤95%. The two levels of temperature “cold” and “warm” correspond to daily temperatures in the ranges: 0 °C < T ≤ 13 °C and 13 °C < T ≤ 28 °C, respectively (Table 3).
Primary infections
The three parameters for primary infection rate, with changing input values according to weather factors, are the rate of primary infections (i.e., the number of primary infection per day, P), the onset date for the first primary infection (in day of the year, OD), and the duration (in number of days) of primary inoculum mobilisation (PD). Each of these three parameters has two levels. P is defined “low” or “high” when its value is 0.02 [N.T−1] or 0.2 [N.T−1], respectively (Gobbin et al. 2005; Rumbou and Gessler 2004). OD is defined “early” or “late”, corresponding to OD = 130 or OD = 160, respectively (Rossi et al. 2008). PD is represented by the two levels “short” and “long”, corresponding to PD = 10 or PD = 60 days, respectively (Rossi et al. 2009).
Secondary infections
The four parameters for secondary infections are the modifiers of the optimum basic infection rate corrected for the removals (RcOpt) i) for temperature (RcT) and ii) for moisture (RcW), iii) for the duration (in days) of the latency period (LP) and iv) for the infectious period (IP).
RcT is defined as “high” or “low” when its value is 1 or 0.8, respectively. High and low RcT correspond to the level of temperature “warm” and “cold”, respectively, and account for the effect of temperature on secondary infections (Caffi et al. 2013; Gregory 1915; Lalancette et al. 1988; Rossi et al. 2013).
RcW has four levels, termed “very low”, “low”, “medium” or “high”, corresponding to 0.1, 0.4, 0.6, and 1, respectively. These values correspond to the level of moisture termed “very dry”, “dry”, “partly wet”, and “wet”, respectively, and account for the effect of moisture on secondary infections (Blaeser and Weltzien 1978; Rossi et al. 2013), via infection efficiency.
LP has two levels termed “short” or “long”, which correspond to 6 and 10 days, respectively, accounting for the effect of temperature and moisture on the duration of latency period (Goidanich 1959; Rossi et al. 2002). IP has two levels termed “long” or “short”, corresponding to 20 and 15 days, respectively, accounting for the effect of temperature and moisture on the duration of infectious period (Hill 1989).
Model evaluation
Evaluation of a simulation model includes model verification and model testing (Teng 1981). Model verification consists in verifying that the model structure and the equations perform calculations as expected. Results from model verification are reported in Bove (2018) and Bove et al. (2020), and allow considering the model as satisfactorily reflecting the hypotheses embedded in its equations. Model testing consists in “testing the agreement between model behaviour and that of the real system” (Teng 1981), and was implemented according to the six considered scenarios of disease conduciveness.
First, the behaviour of the real system was defined with respect to potential epidemics on both leaves and clusters for each of the six scenarios of disease conduciveness. The patterns of the six potential epidemics represent the expected outcomes of the six scenarios, and were derived from disease progress curves on leaves, and disease intensity on clusters reported in the literature (Carisse 2016; Dagostin et al. 2011; Dalla Marta et al. 2005; Gessler and Blaise 1992; Jermini et al. 2001; Savary et al. 2009). These epidemics were monitored under a range of environment in vineyards with susceptible varieties which were not protected against downy mildew. The environmental (weather) conditions prevailing in the vineyards where the epidemics were monitored was characterized together with the patterns of downy mildew epidemics.
Second, a simulation experiment was conducted, whereby the model was run using the sets of parameters corresponding to the six scenarios of disease conduciveness, which allowed generating simulated potential epidemics on leaves and on clusters. For each simulation, parameter values were set according to Table 2. The simulated epidemics were then compared to the expected overall patterns of the pathosystem derived from the literature.
Results
Anticipated disease progress curves of potential downy mildew epidemics
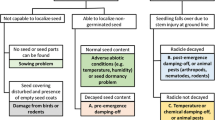
Potential downy mildew epidemics, represented by disease progress curves on leaves and number of infected clusters, are shown in Fig. 1. Six different environmental scenarios prevailing throughout whole grapevine growing seasons were thus associated with distinct disease progress patterns, based on the literature on grape downy mildew.
Expected outcomes of six scenarios of disease conduciveness in downy mildew of grapevine based on literature at fixed levels of primary inoculum. Scenarios include the effect of the temperature (T) and moisture (W) on (1) the number of primary infections per day, (2) the duration of primary inoculum mobilisation, (3) the onset date of primary infections, (4) the durations of the latency and infectious periods, (5) the basic infection rate corrected for the removals Rc. Scenarios do not include the amount of primary inoculum, which is assumed fixed in the considered system
Potential epidemics on leaves in a “wet and warm” climate scenario (Fig. 1, Scenario 1) are expected to be strong, since temperature and moisture are highly favourable for the mobilisation of both primary and secondary inoculum and for the occurrence of both primary and secondary infections. In these conditions, a very strong level of infection on clusters is expected.
Epidemics on foliage are predicted to be strong in the “wet and cold” and “partly wet and warm” (Fig. 1, Scenario 2 and Scenario 3) scenarios too, with some limits to the full intensification of the disease, resulting in a delay of the epidemic and in lower severity on foliage, compared to the first scenario (“wet and warm”, Scenario 1). Both scenarios 2 and 3 are not, however, assumed to be limiting for the disease on clusters, resulting in a very high number of diseased clusters (Fig. 1).
The fourth scenario (Fig. 1, Scenario 4, “partly wet and cold”) is assumed to limit the development of the epidemic: under “partly wet and cold” conditions, the potential s-shape of downy mildew progress curve on the foliage is compromised. Under this scenario, a small amount of primary inoculum is assumed to be mobilised, thus disease progress is delayed. Furthermore, a strong reduction of disease severity on leaves is expected, reflecting a very limited number of secondary infections, and thus, a lower slope of disease progress on the foliage. As a result, few clusters, if any, are expected to be diseased.
Scarce rain, low moisture and low temperature are unfavourable to the development of downy mildew epidemics. Scenarios 5 (“dry and warm”) and 6 (“very dry and warm”) are therefore considered the less favourable to downy mildew. Scenarios 5 and 6 (Fig. 1) are anticipated to be associated with strong suppressions of the epidemic on foliage and clusters.
Simulation experiment and comparison of scenarios
Model simulations in each of the six scenarios are reported in Fig. 2 in three panels: the overall dynamics of (canopy) sites, including healthy (HS), latent (L), infectious (I), removed (R) sites, along with the healthy senesced (SEN) and removed senesced (SENR) sites (Fig. 2, Panel a); the detailed dynamics of infected sites, including latent (L), infectious (I), removed (R), and removed senesced (SENR) sites (Fig. 2, Panel b); and the dynamics of disease on clusters, with healthy (HC) and diseased (DC) clusters, along with total diseased (canopy) sites (D = L + I + R) and infectious (I) sites (Fig. 2, Panel c).
Simulation outputs for 6 scenarios of disease conduciveness. a Dynamics of healthy (HS), latent (L), infectious (I), removed (R), healthy senesced (SEN), and removed senesced (SENR) sites. b Detail of the dynamics of diseased sites: latent, infectious, removed, removed senesced, and total diseased sites on foliage (D). c Dynamics of healthy and diseased clusters in relation with disease on the foliage: healthy (HC) and diseased (DC) clusters, infectious and total diseased sites on foliage. The values of parameters used in each scenario are shown in Table 2. Horizontal axis: time (days) from bud break (day = 0). Vertical left axis: number of sites; the maximum values for graphs a and c represents Smax, the total number of sites that the system can support; graph b is rescaled to show the disease dynamics in detail; vertical right axis (graph c): number of clusters
In scenario 1, the first primary infection occurs at DOS 40. The maximum number of diseased sites on foliage (D = 6600 sites) is reached at DOS 160 (Fig. 2, Scenario 1, Panel b). A sharp decrease of disease on the foliage (Fig. 2, Panel b, D) towards the end of the epidemic results from the outflow of sites from R to SENR, reflecting disease-induced senescence. Infection on clusters starts at DOS 62 and reaches its maximum (20 diseased clusters) at DOS 123. All 20 clusters in the system (one grapevine plant) are infected at the end of the epidemic (Fig. 2, Scenario 1, panel c, DC). In this simulation, the first infectious site appears at DOS 48. When disease on clusters reaches its maximum, there are 900 infectious sites (I) and 1700 diseased sites (L + I + R), over a carrying capacity of 10,000 sites.
In scenario 2, the maximum number of diseased sites on foliage is approximately 1000, and is reached at the end of the simulation (DOS 200, Fig. 2, Scenario 2, Panels a, b). The epidemic is delayed in comparison with the previous, more favourable scenario. Infection on clusters starts at DOS 63 and reaches its maximum (17 diseased clusters out of 20) at DOS 200, i.e., at the end of the simulation. Disease progress on clusters (Fig. 2, Scenario 2, Panel c) is reduced compared to scenario 1 for two reasons: first, a slower initial progress (reflecting lower disease on the canopy; Fig. 2, Panels a, b), and second, simulated ontogenic resistance in clusters. The change in slope of the disease progress curve on clusters (Fig. 2, Scenario 2, Panel c) reflects the switch of development stage from DVS 1 to DVS 2 at DOS 140. Under DVS 2, the susceptibility of clusters to downy mildew is reduced, according to the CCS, as hypothesised in the model.
In Scenario 3, the maximum number of diseased sites on foliage is 1500 sites (Fig. 2, Scenario 3, Panels a,b). The disease incidence on clusters reaches its maximum (20 diseased clusters out of 20) at DOS 154 (Fig. 2, Scenario 3, panel c).
In Scenario 4, the maximum number of diseased sites is 6 out of 10,000 sites (Fig. 2, Scenario 4, Panels a,b). The number of diseased clusters is very small: only 1 cluster over 20 is infected (Fig. 2, Scenario 4, Panel c).
Lastly, scenarios 5 and 6 show negligible disease on leaves and no disease on clusters is simulated (Fig. 2, Panels a,b,c).
Overall, this simulation experiment shows that the most favourable scenario of disease conduciveness is the first one (Fig. 2, scenario 1, Panels a,b,c). However, disease on foliage never reaches its carrying capacity of 10,000 sites (Fig. 2). As shown in Fig. 2 (Panel b, curve I, and Panel c, curve DC), a small fraction of disease on foliage is sufficient to cause a large infection on clusters. From a crop growth standpoint (Fig. 2, Panel a) the total number of sites (D + H + SENR), TOTSITES, reaches the maximum (10,000 = Smax) at DOS 99 in each scenario. This is because the scenarios are different in their effects on disease conduciveness (Table 2), and are assumed to not affect crop growth, crop development, and physiological senescence.
Discussion
The work reported here first provides a quantitative synthesis of knowledge on the effect of weather factors on epidemiological processes involved in grape downy mildew. This represents a further step to the quantitative synthesis of the knowledge on grape downy mildew at the process and at the epidemic levels that preceded this work (Bove, 2018; Bove et al., 2020). The present work focuses on potential disease epidemics (Savary et al. 2018), i.e., epidemics which take place in the absence of any attempt of control (host plant resistance and chemicals). The present work enables defining six weather-based scenarios which capture a range of environmental conditions under which potential epidemics of grape downy mildew may develop. A synthesis of the literature, conducted at the epidemic level, then leads to identify patterns of epidemics under these environmental scenarios (Fig. 1). Our process-based simulation model was then tested when simulated outputs were compared with the anticipated, knowledge-based, behaviour of the modelled system in the six identified scenarios (Fig. 2).
The six scenarios of disease conduciveness defined in this study (Table 2) capture the strong influence of weather on epidemics of downy mildew of grapevine (Rossi et al. 2005), which can be summarised as follows. Temperature, rain and wetness play a critical role in several phases of the life cycle of P. viticola, resulting into strong effects on the epidemic. The first scenario (wet and warm) is the most favourable. These conditions are suitable for a strong mobilisation of primary inoculum and enable multiple secondary infections (Gessler et al. 2011; Rossi et al. 2013). In the second scenario (wet and cold), moisture, which is the strongest limiting factor for the mobilisation of primary inoculum, especially in the form of rainfall events (Galbiati and Longhin 1984; Rossi and Caffi 2012), allows the occurrence of primary infections and some secondary infections. However, low temperatures result in a slower germination of primary inoculum (Rossi et al. 2008), and are less favourable for secondary infections development, since the optimum is about 20 °C (Blaeser and Weltzien 1978; Gessler et al. 2011). The third scenario (partly wet and warm) is characterised by sufficient moisture for the mobilisation of primary inoculum, even if the mobilisation is slower than in the first scenario. In the fourth scenario (cold and partly wet), low temperatures limit the occurrence of both primary and secondary infections (Gessler et al. 2011; Kennelly et al. 2007). However, moisture is sufficient to enable a low epidemic. The fifth scenario (dry and warm), is strongly limiting (low moisture) for the mobilisation of primary inoculum and for the occurrence of sporulation. The sixth scenario (very dry and warm) is the most unfavourable for downy mildew epidemic development.
The behaviour of (uncontrolled) potential epidemics (occurring on susceptible varieties, without fungicide application) of downy mildew of grapevine in six scenarios of disease conduciveness was derived from our knowledge from the literature and represented the first step to test the model. A visual comparison of the simulated output of the model and the anticipated patterns suggests that the model accurately simulate epidemics according to climate scenarios. Simulated outputs derived from the simulation in Scenario 1 (disease progress on foliage and clusters) were congruent with the epidemic analysis (Fig. 1), in which a very high level of disease on both foliage and clusters was assumed. Simulations in Scenario 2 were also congruent with anticipated patterns, in which a high level of disease on foliage and cluster was expected. A simulated delay of the epidemic was anticipated in the potential epidemic analysis. However, the shape of the disease progress curve on foliage did not correspond to the predicted s-shape, because of the strong delay in the epidemic. Scenario 2 mainly differs from Scenario 1 in having a lower rate of infection, RI, because of lower RcT, longer LP and shorter IP. Both RcT and LP contribute to delay the epidemic, as observed in the simulation (Fig. 2, Scenario 2). Simulation in Scenario 3 (Fig. 2, Scenario 3) is similar to the anticipated epidemic (Fig. 1, Scenario 3). The simulated number of diseased clusters in this scenario is very high. The rate of infection is lower than in Scenario 2, because of a lower RcW (Table 2). This explains why the simulated epidemic on foliage in Scenario 3 is slower than in the first scenario and the slope of the curve is delayed (Fig. 2, Scenario 3). Negligible epidemics on foliage and clusters are simulated in Scenarios 4, 5 and 6 (Fig. 2). Simulated epidemics on foliage and clusters in Scenario 5 and 6 were null, as expected from the analysis of potential epidemics (Fig. 1, Scenarios 5 and 6, infections on clusters). However, simulation in Scenario 4 was expected from the potential analysis to produce some disease on foliage and clusters (Fig. 1, Scenario 4), because of sufficient moisture for the mobilisation of primary inoculum and occurrence of secondary infections (Table 2). In this case, the anticipated epidemics were overestimated. The low amount of primary infections and the long duration of LP characterizing Scenario 4 have a strong effect on delaying the simulated epidemic. Moreover, the low values of the parameters RcT and RcW, contributed to suppress the rate of infection.
The model discussed in this work performs as intended, simulating potential dual epidemics of downy mildew across different sets of environmental conditions driving the disease. In all of the six scenarios accounting for changes in environmental conditions, the model was able to mobilise existing quantitative information on downy mildew of grapevine and to generate outputs that are congruent with estimated patterns of potential epidemics. Model testing, in which simulated epidemics were compared to anticipated epidemics on both foliage and clusters, lead us to conclude that this modelling structure produces accurate and robust outputs. This study indicates that the model is a reliable tool for simulating potential epidemics of downy mildew of grapevine in a scenario analysis. This can have many applications, such as the understanding of the behaviour of the pathosystem under climate change or when partial host plant resistance is involved (Bove 2018).
References
Bernizzoni, F., Gatti, M., Civardi, S., & Poni, S. (2009). Long-term performance of Barbera grown under different training systems and within-row vine spacings. American Journal of Enology and Viticulture, 60(3), 339–348.
Blaeser, M., & Weltzien, H. C. (1978). The importance of sporulation, dispersal, and germination of sporangia of Plasmopara viticola. Journal of Plant Disease and Protection, 3, 155–161.
Bove, F. (2018). A modelling framework for grapevine downy mildew epidemics incorporating foliage-cluster relationships and host plant resistance. Piacenza: Università Cattolica del Sacro Cuore Retrieved from http://hdl.handle.net/10280/57899.
Bove, F., Savary, S., Willocquet, L., & Rossi, V. (2020). Designing a modelling structure for the grapevine downy mildew pathosystem. European Journal of Plant Pathology, 157, 251–268. https://doi.org/10.1007/s10658-020-01974-2.
Caffi, T., Gilardi, G., Monchiero, M., & Rossi, V. (2013). Production and release of asexual sporangia in Plasmopara viticola. Phytopathology, 103(1), 64–73. https://doi.org/10.1094/PHYTO-04-12-0082-R.
Carisse, O. (2016). Development of grape downy mildew (Plasmopara viticola) under northern viticulture conditions: Influence of fall disease incidence. European Journal of Plant Pathology, 144(4), 773–783. https://doi.org/10.1007/s10658-015-0748-y.
Cola, G., Mariani, L., Salinari, F., Civardi, S., Bernizzoni, F., Gatti, M., & Poni, S. (2014). Description and testing of a weather-based model for predicting phenology, canopy development and source-sink balance in Vitis vinifera L. cv. Barbera. Agricultural and Forest Meteorology, 184, 117–136. https://doi.org/10.1016/j.agrformet.2013.09.008.
Dagostin, S., Schärer, H. J., Pertot, I., & Tamm, L. (2011). Are there alternatives to copper for controlling grapevine downy mildew in organic viticulture? Crop Protection, 30(7), 776–788. https://doi.org/10.1016/j.cropro.2011.02.031.
Dalla Marta, A., Magarey, R. D., & Orlandini, S. (2005). Modelling leaf wetness duration and downy mildew simulation on grapevine in Italy. Agricultural and Forest Meteorology, 132(1–2), 84–95. https://doi.org/10.1016/j.agrformet.2005.07.003.
Galbiati, C., & Longhin, G. (1984). Indagini sulla formazione e sulla germinazione delle oospore di Plasmopara viticola. Rivista di Patologia Vegetale, 20(2), 66–80.
Galet, P. (1977). Les Maladies et les Parasites de la Vigne (Vol. 1). Montpellier: Imprimerie du Paysan du Midi.
Gatti, M., Pirez, F. J., Chiari, G., Tombesi, S., Palliotti, A., Merli, M. C., & Poni, S. (2016). Phenology, canopy aging and seasonal carbon balance as related to delayed winter pruning of Vitis vinifera L. cv. sangiovese grapevines. Frontiers in Plant Science, 7, 659. https://doi.org/10.3389/fpls.2016.00659.
Gessler, C., & Blaise, P. (1992). An extended progeny/parent ratio model II. Application to experimental data. Journal of Phytopathology, 134(1), 53–62. https://doi.org/10.1111/j.1439-0434.1992.tb01211.x.
Gessler, C., Pertot, I., & Perazzolli, M. (2011). Plasmopara viticola: A review of knowledge on downy mildew of grapevine and effective disease management. Phytopathologia Mediterranea, 50(1), 3–44.
Gobbin, D., Jermini, M., Loskill, B., Pertot, I., Raynal, M., & Gessler, C. (2005). Importance of secondary inoculum of Plasmopara viticola to epidemics of grapevine downy mildew. Plant Pathology, 54(4), 522–534. https://doi.org/10.1111/j.1365-3059.2005.01208.x.
Goidanich, G. (1959). Manuale di patologia vegetale - v. 1–4. Bologna: Edizioni Agricole.
Gregory, C. T. (1915). Studies on Plasmopara viticola (Downey mildew of grapes). Proceedings of the International Congress of Viticulture (San Francisco, CA), 126–150.
Gutierrez, A. P., Williams, D. W., & Kido, H. (1985). A model of grape growth and development: The mathematical structure and biological considerations. Crop Science, 25(5), 721–728. https://doi.org/10.2135/cropsci1985.0011183x002500050001x.
Hill, G. K. (1989). Effect of temperature on sporulation efficiency of oilspots caused by Plasmopara viticola (Berk. & Curt. ex De Bary) Berl. & De Toni in the vineyard. Viticultural and Enological Sciences, 44, 86–90.
Jermini, M., Blaise, P., & Gessler, C. (2001). Quantification of the influence of Plasmopara viticola on Vitis vinifera as a basis for the optimisation of the control. In Integrated Control in Viticulture IOBC wprs Bulletin (Vol. 24, pp. 37–44). https://doi.org/10.1016/0167-8809(89)90025-x.
Jermini, M., Blaise, P., & Gessler, C. (2010a). Quantification of the influence of the downy mildew (Plasmopara viticola) epidemics on the compensatory capacities of Vitis vinifera “merlot” to limit the qualitative yield damage. Vitis, 49, 153–160.
Jermini, M., Blaise, P., & Gessler, C. (2010b). Response of “merlot” (Vitis vinifera) grapevine to defoliation caused by downy mildew (Plasmopara viticola) during the following growing season. Vitis, 49, 161–166.
Jermini, Mauro, Carrera, E., Dietrich, R., & Blaise, P. (2000). Impact of downy mildew epidemics on grapevine development in the following season. In Integrated Control in Viticulture IOBC wprs Bulletin (Vol. 23, pp. 9–12).
Kennelly, M. M., Gadoury, D. M., Wilcox, W. F., Magarey, P. A., & Seem, R. C. (2005). Seasonal development of ontogenic resistance to downy mildew in grape berries and rachises. Phytopathology, 95(12), 1445–1452. https://doi.org/10.1094/PHYTO-95-1445.
Kennelly, M. M., Gadoury, D. M., Wilcox, W. F., Magarey, P. A., & Seem, R. C. (2007). Primary infection, lesion productivity, and survival of sporangia in the grapevine downy mildew pathogen Plasmopara viticola. Phytopathology, 97(4), 512–522. https://doi.org/10.1094/PHYTO-97-4-0512.
Lalancette, N. (1987). Estimating infection efficiency of Plasmopara viticola on grape. Plant Disease, 71(11), 981. https://doi.org/10.1094/pd-71-0981.
Lalancette, N. (1988). Development of an infection efficiency model for Plasmopara viticola on American grape based on temperature and duration of leaf wetness. Phytopathology, 78(6), 794–800. https://doi.org/10.1094/phyto-78-794.
Lalancette, N., Madden, L. V., & Ellis, M. A. (1988). A quantitative model for describing the sporulation of Plasmopara viticola on grape leaves. Phytopathology, 78(10), 1316–1321. https://doi.org/10.1094/phyto-78-1316.
Loomis, R. S., & Adams, S. S. (1983). Integrative analyses of host-pathogen relations. Annual Review of Phytopathology, 21(1), 341–362. https://doi.org/10.1146/annurev.py.21.090183.002013.
Müller, K., & Sleumer, H. (1934). Biologische Untersuchungen über die Peronosporakrankheit des Weinstockes mit besonderer Berücksichtigung ihrer Bekämpfung nach der Inkubationskalendermethode. Landwirtschaftliche Jahrbucher, 79(4), 509–576.
Poni, S., Palliotti, A., & Bernizzoni, F. (2006). Calibration and evaluation of a STELLA software-based daily CO2 balance model in Vitis vinifera L. Journal of the American Society for Horticultural Science, 131(2), 273–283.
Reuveni, M. (1998). Relationships between leaf age, peroxidase and β-1, 3-GIucanase activity, and resistance to downy mildew in grapevines. Journal of Phytopathology, 146(10), 525–530.
Rossi, V., Giosue, S., Girometta, B., & Bugiani, R. (2002). Influence of climatic conditions on primary infections caused by Plasmopara viticola in Emilia-Romagna [Vitis vinifera L.]. In Atti delle Giornate Fitopatologiche.
Rossi, V., Giosuè, S., & Caffi, T. (2009). Modelling the dynamics of infections caused by sexual and asexual spores during Plasmopara Viticola epidemics. Journal of Plant Pathology, 91(3), 615–627. https://doi.org/10.1177/0191453704045756.
Rossi, V., & Caffi, T. (2007). Effect of water on germination of Plasmopara viticola oospores. Plant pathology, 56(6), 957–966. https://doi.org/10.1111/j.1365-3059.2007.01685.x
Rossi, V., & Caffi, T. (2012). The role of rain in dispersal of the primary inoculum of Plasmopara viticola. Phytopathology, 102(2), 158–165. https://doi.org/10.1094/PHYTO-08-11-0223.
Rossi, V., Caffi, T., Giosuè, S., & Bugiani, R. (2008). A mechanistic model simulating primary infections of downy mildew in grapevine. Ecological Modelling, 212(3–4), 480–491. https://doi.org/10.1016/j.ecolmodel.2007.10.046.
Rossi, V., Caffi, T., & Gobbin, D. (2013). Contribution of molecular studies to botanical epidemiology and disease modelling: Grapevine downy mildew as a case-study. European Journal of Plant Pathology, 135(4), 641–654. https://doi.org/10.1007/s10658-012-0114-2.
Rossi, V., Caffi, T., Melandri, M., & Pradolesi, G. (2005). Aggiornamenti sulla peronospora della vite. Agronomica, 2, 38–56 http://hdl.handle.net/10807/45995%0A.
Rumbou, A., & Gessler, C. (2004). Genetic dissection of Plasmopara viticola population from a Greek vineyard in two consecutive years. European Journal of Plant Pathology, 110(4), 379–392. https://doi.org/10.1023/B:EJPP.0000021061.38154.22.
Rumbou, A., & Gessler, C. (2006). Particular structure of Plasmopara viticola populations evolved under Greek island conditions. Phytopathology, 96(5), 501–509. https://doi.org/10.1094/PHYTO-96-0501.
Savary, S., Delbac, L., Rochas, A., Taisant, G., & Willocquet, L. (2009). Analysis of nonlinear relationships in dual epidemics, and its application to the management of grapevine downy and powdery mildews. Phytopathology, 99(8), 930–942. https://doi.org/10.1094/PHYTO-99-8-0930.
Savary, S., Nelson, A. D., Djurle, A., Esker, P. D., Sparks, A., Amorim, L., Bergamin Filho, A., Caffi, T., Castilla, N., Garrett, K., McRoberts, N., Rossi, V., Yuen, J., & Willocquet, L. (2018). Concepts, approaches, and avenues for modelling crop health and crop losses. European Journal of Agronomy, 100, 4–18. https://doi.org/10.1016/j.eja.2018.04.003.
Savary, S., & Willocquet, L. (2014). Simulation modeling in botanical epidemiology and crop loss analysis. The Plant Health Instructor. APSnet Education Center. https://doi.org/10.1094/phi-a-2014-0314-01.
Sun, P., & Zeng, S. (1994). On the measurement of the corrected basic infection rate. Zeitschrift fur Pflanzenkrankheiten und Pflanzenschutz, 101(3), 297–302.
Teng, P. S., Blackie, M. J., & Close, R. C. (1977). A simulation analysis of crop yield loss due to rust disease. Agricultural Systems, 2(3), 189–198. https://doi.org/10.1016/0308-521X(77)90004-X.
Teng, P. S. (1981). Validation of computer models of plant disease epidemics: A review of philosophy and methodology. Zeitschrift für Pflanzenkrankheiten und Pflanzenschutz/Journal of Plant Diseases and Protection, 88(1), 49–63 http://www.jstor.org/stable/43214777.
Vanderplank, J. E. (1963). Plant diseases: Epidemics and control. New York: Academic Press Inc..
Vanderplank, J. E. (1975). Principles of plant infection. New York: Academic Press Inc..
Wermelinger, B., & Koblet, W. (1990). Seasonal growth and nitrogen distribution in grapevine leaves, shoots and grapes. Vitis, 29(1), 15–26.
Willocquet, L., Aubertot, J. N., Lebard, S., Robert, C., Lannou, C., & Savary, S. (2008). Simulating multiple pest damage in varying winter wheat production situations. Field Crops Research, 107(1), 12–28. https://doi.org/10.1016/j.fcr.2007.12.013.
Winkler, A. J., Cook, J. A., Kliewer, W. M., & Lider, L. A. (1974). The physiology of the vine. In General viticulture (pp. 90–109). Berkeley: University of California Press.
Zadoks, J. C., & Schein, R. D. (1979). Epidemiology and plant disease management. New York: Oxford Univ. Press.
Acknowledgments
This study was supported by the Doctoral School on the Agro-Food System (Agrisystem) of the Università Cattolica del Sacro Cuore (Italy).
Funding
Open access funding provided by Università Cattolica del Sacro Cuore within the CRUI-CARE Agreement.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
The Authors declare that the present manuscript complies with the Ethical Rules of good scientific practice applicable for the European Journal of Plant Pathology.
Conflict of interest
The authors declare that they have no conflict of interest. All authors are informed and agree on the publication of the manuscript.
Research involving human participants and/or animals
Not applicable, the research does not involve humans or animals.
Informed consent
Not applicable, the research does not involve human participants.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Bove, F., Savary, S., Willocquet, L. et al. Simulation of potential epidemics of downy mildew of grapevine in different scenarios of disease conduciveness. Eur J Plant Pathol 158, 599–614 (2020). https://doi.org/10.1007/s10658-020-02085-8
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10658-020-02085-8