Cytokinesis-Block Micronucleus Cytome Assay Evolution into a More Comprehensive Method to Measure Chromosomal Instability
Abstract
:1. Background
2. The Origin of Micronuclei
“Nonetheless, I regard irregularities of mitosis as the usual way in which a nucleus of inappropriate composition is generated. I would like to say a few more words about this. So-called asymmetrical mitoses, which do not appear to be rare, may have much the same effect as multipolar mitoses. Asymmetrical mitoses, as the analysis of suitable material has shown, arise when the fibres associated with one of the two centrospheres do not form an attachment to all the chromosomes. The chromosomes that are attached to only one set of fibres—or, if division of the chromosome has already begun, the two unseparated daughter chromosomes—then pass to one of the daughter cells, whereas the other daughter cell remains devoid of these elements. It is precisely these aberrations that can easily lead to a marked numerical excess of one type of chromosome relative to the others, if that is what is required.”[5]
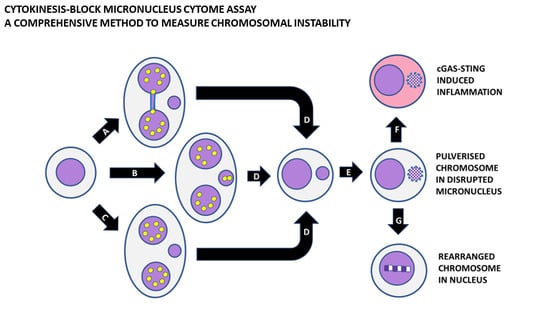
3. The Cytokinesis-Block Micronucleus Assay
4. The CBMN Cytome Assay to Measure Chromosomal Instability
5. Use of Molecular Probes in the CBMN Cytome Assay to Understand the Mechanisms
6. Other Emerging Biomarkers in the CBMN Cytome Assay
7. Shattering of Chromosomes in MNi and Their Subsequent Hypermutation and Inflammation
8. Conclusions and Future Directions
- (1)
- What are the preventable causes of MNi, NPBs, and NBUD formation?
- (2)
- What are the most important genetic causes of MNi, NPBs, and NBUD formation?
- (3)
- Are cGAS-STING induction and chromoanagenesis mutually exclusive or do they occur one after the other in the same cell?
- (4)
- What are the molecular differences between MNi that remain extranuclear and trigger cGAS-STING versus MNi that result in chromothripsis followed by nuclear reintegration and chromoanagenesis?
- (5)
- What are the best probes to detect chromosome pulverisation in MNi, induction of cGAS-STING by MNi, and chromoanagenesis of a pulverised chromosome within a nucleus?
- (6)
- Is it possible to further enhance the lymphocyte CBMN cytome assay by automation so that it becomes practical to use it as a high-content molecular cytogenetic assay using multiple probes simultaneously not only as a research tool to study mechanisms but also in routine clinical diagnostics of healthy aging?
Funding
Conflicts of Interest
Abbreviations
| BN | binucleated |
| CBMN | cytokinesis-block micronucleus |
| cGAS | cyclic GMP-AMP synthase |
| CIN | chromosomal instability |
| FISH | fluorescence in situ hybridisation |
| MN | micronucleus |
| MNi | micronuclei |
| NBUDs | nuclear buds |
| NPBs | nucleoplasmic bridges |
| PNA | peptide nucleic acid |
| RBCs | red blood cells |
| STING | Stimulator of Interferon Genes |
| UFBs | ultra-fine bridges |
References
- Daughtry, B.L.; Chavez, S.L. Chromosomal instability in mammalian pre-implantation embryos: Potential causes, detection methods, and clinical consequences. Cell Tissue Res. 2016, 363, 201–225. [Google Scholar] [CrossRef] [PubMed]
- Bakhoum, S.F.; Cantley, L.C. The Multifaceted Role of Chromosomal Instability in Cancer and Its Microenvironment. Cell 2018, 174, 1347–1360. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Barroso-Vilares, M.; Logarinho, E. Chromosomal instability and pro-inflammatory response in aging. Mech. Ageing Dev. 2019, 182, 111118. [Google Scholar] [CrossRef] [PubMed]
- Rao, C.V.; Asch, A.S.; Yamada, H.Y. Emerging links among Chromosome Instability (CIN), cancer, and aging. Mol. Carcinog. 2017, 56, 791–803. [Google Scholar] [CrossRef]
- Boveri, T. Concerning the origin of malignant tumours by Theodor Boveri. Translated and annotated by Henry Harris. J. Cell Sci. 2008, 121 (Suppl. 1), 1–84. [Google Scholar] [CrossRef]
- Sears, D.A.; Udden, M.M. Howell-Jolly bodies: A brief historical review. Am. J. Med. Sci. 2012, 343, 407–409. [Google Scholar] [CrossRef]
- Bain, B.J. Prominent Howell-Jolly bodies when megaloblastic anemia develops in a hyposplenic patient. Am. J. Hematol. 2014, 89, 852. [Google Scholar] [CrossRef] [Green Version]
- Everson, R.B.; Wehr, C.M.; Erexson, G.L.; MacGregor, J.T. Association of marginal folate depletion with increased human chromosomal damage in vivo: Demonstration by analysis of micronucleated erythrocytes. J. Natl. Cancer Inst. 1988, 80, 525–529. [Google Scholar] [CrossRef]
- Dawson, D.W.; Bury, H.P. The significance of Howell-Jolly bodies and giant metamyelocytes in marrow smears. J. Clin. Pathol. 1961, 14, 374–380. [Google Scholar] [CrossRef] [Green Version]
- Heddle, J.A.; Fenech, M.; Hayashi, M.; MacGregor, J.T. Reflections on the development of micronucleus assays. Mutagenesis 2011, 26, 3–10. [Google Scholar] [CrossRef]
- Hayashi, M.; Tice, R.R.; MacGregor, J.T.; Anderson, D.; Blakey, D.H.; Kirsh-Volders, M.; Oleson, F.B., Jr.; Pacchierotti, F.; Romagna, F.; Shimada, H. In vivo rodent erythrocyte micronucleus assay. Mutat. Res. 1994, 312, 293–304. [Google Scholar] [CrossRef]
- Odagiri, Y.; Takemoto, K.; Fenech, M. Micronucleus induction in cytokinesis-blocked mouse bone marrow cells in vitro following in vivo exposure to X-irradiation and cyclophosphamide. Environ. Mol. Mutagen 1994, 24, 61–67. [Google Scholar] [CrossRef] [PubMed]
- Fenech, M. Cytokinesis-block micronucleus cytome assay. Nat. Protoc. 2007, 2, 1084–1104. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fenech, M.; Perepetskaya, G.; Mikhalevich, L. A more comprehensive application of the micronucleus technique for biomonitoring of genetic damage rates in human populations—Experiences from the Chernobyl catastrophe. Environ. Mol. Mutagen 1997, 30, 112–118. [Google Scholar] [CrossRef]
- Fenech, M.; Knasmueller, S.; Bolognesi, C.; Bonassi, S.; Holland, N.; Migliore, L.; Palitti, F.; Natarajan, A.T.; Kirsch-Volders, M. Molecular mechanisms by which in vivo exposure to exogenous chemical genotoxic agents can lead to micronucleus formation in lymphocytes in vivo and ex vivo in humans. Mutat. Res. 2016, 770 Pt A, 12–25. [Google Scholar] [CrossRef]
- Kirsch-Volders, M.; Fenech, M.; Bolognesi, C. Validity of the Lymphocyte Cytokinesis-Block Micronucleus Assay (L-CBMN) as biomarker for human exposure to chemicals with different modes of action: A synthesis of systematic reviews. Mutat. Res. Genet. Toxicol. Environ. Mutagen 2018, 836 Pt A, 47–52. [Google Scholar] [CrossRef]
- Kirsch-Volders, M.; Fenech, M. Micronucleus Assays with Human Lymphocytes for In Vitro Genetic Toxicology Testing. In The Micronucleus Assay in Toxicology; Knasmüller, S., Fenech, M., Eds.; The Royal Society of Chemistry: London, UK, 2019; pp. 157–168. [Google Scholar]
- Kirsch-Volders, M.; Decordier, I.; Elhajouji, A.; Plas, G.; Aardema, M.J.; Fenech, M. In vitro genotoxicity testing using the micronucleus assay in cell lines, human lymphocytes and 3D human skin models. Mutagenesis 2011, 26, 177–184. [Google Scholar] [CrossRef] [PubMed]
- OECD. Test No. 487: In Vitro Mammalian Cell Micronucleus Test; OECD Publishing: Paris, France, 2014. [Google Scholar] [CrossRef]
- Fenech, M. The lymphocyte cytokinesis-block micronucleus cytome assay and its application in radiation biodosimetry. Health Phys. 2010, 98, 234–243. [Google Scholar] [CrossRef]
- International Atomic Energy Agency. Cytogenetic Dosimetry: Applications in Preparedness for and Response to Radiation Emergencies; International Atomic Energy Agency: Vienna, Austria, 2011. [Google Scholar]
- ISO 17099. Radiological Protection—Performance Criteria for Laboratories Using the Cytokinesis Block Micronucleus (CBMN) Assay in Peripheral Blood Lymphocytes for Biological Dosimetry. 2014. Available online: https://www.iso.org/standard/59141.html (accessed on 30 November 2014).
- Fenech, M.; Kirsch-Volders, M.; Natarajan, A.T.; Surralles, J.; Crott, J.W.; Parry, J.; Norppa, H.; Eastmond, D.A.; Tucker, J.D.; Thomas, P. Molecular mechanisms of micronucleus, nucleoplasmic bridge and nuclear bud formation in mammalian and human cells. Mutagenesis 2011, 26, 125–132. [Google Scholar] [CrossRef] [Green Version]
- Fenech, M.F. Dietary reference values of individual micronutrients and nutriomes for genome damage prevention: Current status and a road map to the future. Am. J. Clin. Nutr. 2010, 91, 1438S–1454S. [Google Scholar] [CrossRef] [Green Version]
- Fenech, M. Perspectives in Nutrigenomics and Nutrigenetics. Sight Life 2015, 29, 64–70. [Google Scholar]
- Ye, C.J.; Sharpe, Z.; Alemara, S.; Mackenzie, S.; Liu, G.; Abdallah, B.; Horne, S.; Regan, S.; Heng, H.H. Micronuclei and Genome Chaos: Changing the System Inheritance. Genes 2019, 1, 366. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fenech, M. A mathematical model of the in vitro micronucleus assay predicts false negative results if micronuclei are not specifically scored in binucleated cells or in cells that have completed one nuclear division. Mutagenesis 2000, 15, 329–336. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fenech, M.; Morley, A.A. Cytokinesis-block micronucleus method in human lymphocytes: Effect of in vivo ageing and low dose X-irradiation. Mutat. Res. 1986, 161, 193–198. [Google Scholar] [CrossRef]
- Fenech, M.; Morley, A.A. Measurement of micronuclei in lymphocytes. Mutat. Res. 1985, 147, 29–36. [Google Scholar] [CrossRef]
- Thomas, P.; Umegaki, K.; Fenech, M. Nucleoplasmic bridges are a sensitive measure of chromosome rearrangement in the cytokinesis-block micronucleus assay. Mutagenesis 2003, 18, 187–194. [Google Scholar] [CrossRef] [Green Version]
- Bull, C.F.; Mayrhofer, G.; Zeegers, D.; Mun, G.L.; Hande, M.P.; Fenech, M.F. Folate deficiency is associated with the formation of complex nuclear anomalies in the cytokinesis-block micronucleus cytome assay. Environ. Mol. Mutagen 2012, 53, 311–323. [Google Scholar] [CrossRef]
- Utani, K.; Okamoto, A.; Shimizu, N. Generation of micronuclei during interphase by coupling between cytoplasmic membrane blebbing and nuclear budding. PLoS ONE 2011, 6, e27233. [Google Scholar] [CrossRef] [Green Version]
- Oobatake, Y.; Shimizu, N. Double-strand breakage in the extrachromosomal double minutes triggers their aggregation in the nucleus, micronucleation, and morphological transformation. Genes Chromosomes Cancer 2020, 59, 133–143. [Google Scholar] [CrossRef]
- Haaf, T.; Raderschall, E.; Reddy, G.; Ward, D.C.; Radding, C.M.; Golub, E.I. Sequestration of mammalian Rad51-recombination protein into micronuclei. J. Cell Biol. 1999, 144, 11–20. [Google Scholar] [CrossRef] [Green Version]
- Fenech, M.; Crott, J.W. Micronuclei, nucleoplasmic bridges and nuclear buds induced in folic acid deficient human lymphocytes-evidence for breakage-fusion-bridge cycles in the cytokinesis-block micronucleus assay. Mutat. Res. 2002, 504, 131–136. [Google Scholar] [CrossRef]
- El-Zein, R.A.; Schabath, M.B.; Etzel, C.J.; Lopez, M.S.; Franklin, J.D.; Spitz, M.R. Cytokinesis-blocked micronucleus assay as a novel biomarker for lung cancer risk. Cancer Res. 2006, 66, 6449–6456. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kirsch-Volders, M.; Bonassi, S.; Knasmueller, S.; Holland, N.; Bolognesi, C.; Fenech, M.F. Commentary: Critical questions, misconceptions and a road map for improving the use of the lymphocyte cytokinesis-block micronucleus assay for in vivo biomonitoring of human exposure to genotoxic chemicals—A HUMN project perspective. Mutat. Res. Rev. Mutat. Res. 2014, 759, 49–58. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rodrigues, M.A.; Beaton-Green, L.A.; Wilkins, R.C.; Fenech, M.F. The potential for complete automated scoring of the cytokinesis block micronucleus cytome assay using imaging flow cytometry. Mutat. Res. Genet. Toxicol. Environ. Mutagen 2018, 836 Pt A, 53–64. [Google Scholar] [CrossRef] [PubMed]
- Decordier, I.; Kirsch-Volders, M. Fluorescence in situ hybridization (FISH) technique for the micronucleus test. Methods Mol. Biol. 2013, 1044, 237–244. [Google Scholar] [CrossRef]
- Fucic, A.; Katic, J.; Fthenou, E.; Kogevinas, M.; Plavec, D.; Koppe, J.; Batinic, D.; Chalkiadaki, G.; Chatzi, L.; Lasan, R.; et al. Increased frequency of micronuclei in mononucleated lymphocytes and cytome analysis in healthy newborns as an early warning biomarkers of possible future health risks. Reprod. Toxicol. 2013, 42, 110–115. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Thomas, P.; Xue, J.; Fenech, M. Folate deficiency induces aneuploidy in human lymphocytes in vitro-evidence using cytokinesis-blocked cells and probes specific for chromosomes 17 and 21. Mutat. Res. 2004, 551, 167–180. [Google Scholar] [CrossRef] [PubMed]
- Lindberg, H.K.; Wang, X.; Järventaus, H.; Falck, G.C.; Norppa, H.; Fenech, M. Origin of nuclear buds and micronuclei in normal and folate-deprived human lymphocytes. Mutat. Res. 2007, 617, 33–45. [Google Scholar] [CrossRef]
- Zaguia, N.; Laplagne, E.; Colicchio, B.; Cariou, O.; Al Jawhari, M.; Heidingsfelder, L.; Hempel, W.M.; Jrad, B.B.H.; Jeandidier, E.; Dieterlen, A.; et al. A new tool for genotoxic risk assessment: Reevaluation of the cytokinesis-block micronucleus assay using semi-automated scoring following telomere and centromere staining. Mutat. Res. 2020, 850–851, 503143. [Google Scholar] [CrossRef]
- Lee, S.L.; Thomas, P.; Fenech, M. Extracellular amyloid β 42 causes necrosis, inhibition of nuclear division, and mitotic disruption under both folate deficient and folate replete conditions as measured by the cytokinesis-block micronucleus cytome assay. Environ. Mol. Mutagen 2014, 55, 1–14. [Google Scholar] [CrossRef]
- Remeseiro, S.; Cuadrado, A.; Carretero, M.; Martínez, P.; Drosopoulos, W.C.; Cañamero, M.; Schildkraut, C.L.; Blasco, M.A.; Losada, A. Cohesin-SA1 deficiency drives aneuploidy and tumourigenesis in mice due to impaired replication of telomeres. EMBO J. 2012, 31, 2076–2089. [Google Scholar] [CrossRef]
- Deardorff, M.A.; Wilde, J.J.; Albrecht, M.; Dickinson, E.; Tennstedt, S.; Braunholz, D.; Mönnich, M.; Yan, Y.; Xu, W.; Gil-Rodríguez, M.C.; et al. RAD21 mutations cause a human cohesinopathy. Am. J. Hum. Genet. 2012, 90, 1014–1027. [Google Scholar] [CrossRef] [Green Version]
- Martin, C.A.; Murray, J.E.; Carroll, P.; Leitch, A.; Mackenzie, K.J.; Halachev, M.; Fetit, A.E.; Keith, C.; Bicknell, L.S.; Fluteau, A.; et al. Mutations in genes encoding condensin complex proteins cause microcephaly through decatenation failure at mitosis. Genes Dev. 2016, 30, 2158–2172. [Google Scholar] [CrossRef] [Green Version]
- Hellmuth, S.; Böttger, F.; Pan, C.; Mann, M.; Stemmann, O. PP2A delays APC/C-dependent degradation of separase-associated but not free securin. EMBO J. 2014, 33, 1134–1147. [Google Scholar] [CrossRef]
- Chan, Y.W.; West, S.C. A new class of ultrafine anaphase bridges generated by homologous recombination. Cell Cycle 2018, 17, 2101–2109. [Google Scholar] [CrossRef] [Green Version]
- Zhang, C.Z.; Spektor, A.; Cornils, H.; Francis, J.M.; Jackson, E.K.; Liu, S.; Meyerson, M.; Pellman, D. Chromothripsis from DNA damage in micronuclei. Nature 2015, 522, 179–184. [Google Scholar] [CrossRef] [Green Version]
- Lewis, C.W.; Golsteyn, R.M. Cancer cells that survive checkpoint adaptation contain micronuclei that harbor damaged DNA. Cell Cycle 2016, 15, 3131–3145. [Google Scholar] [CrossRef] [PubMed]
- Holland, A.J.; Cleveland, D.W. Chromoanagenesis and cancer: Mechanisms and consequences of localized, complex chromosomal rearrangements. Nat. Med. 2012, 18, 1630–1638. [Google Scholar] [CrossRef] [Green Version]
- Fukami, M.; Kurahashi, H. Clinical Consequences of Chromothripsis and Other Catastrophic Cellular Events. Methods Mol. Biol. 2018, 176, 21–33. [Google Scholar] [CrossRef]
- Liu, S.; Kwon, M.; Mannino, M.; Yang, N.; Renda, F.; Khodjakov, A.; Pellman, D. Nuclear envelope assembly defects link mitotic errors to chromothripsis. Nature 2018, 561, 551–555. [Google Scholar] [CrossRef] [PubMed]
- Terzoudi, G.I.; Karakosta, M.; Pantelias, A.; Hatzi, V.I.; Karachristou, I.; Pantelias, G. Stress induced by premature chromatin condensation triggers chromosome shattering and chromothripsis at DNA sites still replicating in micronuclei or multinucleate cells when primary nuclei enter mitosis. Mutat. Res. Genet. Toxicol. Environ. Mutagen 2015, 793, 185–198. [Google Scholar] [CrossRef] [PubMed]
- Pantelias, A.; Karachristou, I.; Georgakilas, A.G.; Terzoudi, G.I. Interphase Cytogenetic Analysis of Micronucleated and Multinucleated Cells Supports the Premature Chromosome Condensation Hypothesis as the Mechanistic Origin of Chromothripsis. Cancers 2019, 11, 1123. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ly, P.; Brunner, S.F.; Shoshani, O.; Kim, D.H.; Lan, W.; Pyntikova, T.; Flanagan, A.M.; Behjati, S.; Page, D.C.; Campbell, P.J.; et al. Chromosome segregation errors generate a diverse spectrum of simple and complex genomic rearrangements. Nat. Genet. 2019, 51, 705–715. [Google Scholar] [CrossRef] [PubMed]
- Ly, P.; Teitz, L.S.; Kim, D.H.; Shoshani, O.; Skaletsky, H.; Fachinetti, D.; Page, D.C.; Cleveland, D.W. Selective Y centromere inactivation triggers chromosome shattering in micronuclei and repair by non-homologous end joining. Nat. Cell Biol. 2017, 19, 68–75. [Google Scholar] [CrossRef] [Green Version]
- Forment, J.V.; Kaidi, A.; Jackson, S.P. Chromothripsis and cancer: Causes and consequences of chromosome shattering. Nat. Rev. Cancer 2012, 12, 663–670. [Google Scholar] [CrossRef]
- Waldron, D. Genome stability: Chromothripsis and micronucleus formation. Nat. Rev. Genet. 2015, 16, 376–377. [Google Scholar] [CrossRef] [PubMed]
- Okamoto, A.; Utani, K.; Shimizu, N. DNA replication occurs in all lamina positive micronuclei, but never in lamina negative micronuclei. Mutagenesis 2012, 27, 323–327. [Google Scholar] [CrossRef] [Green Version]
- Hatch, E.M.; Fischer, A.H.; Deerinck, T.J.; Hetzer, M.W. Catastrophic nuclear envelope collapse in cancer cell micronuclei. Cell 2013, 154, 47–60. [Google Scholar] [CrossRef] [Green Version]
- Guo, X.; Dai, X.; Wu, X.; Zhou, T.; Ni, J.; Xue, J.; Wang, X. Understanding the birth of rupture-prone and irreparable micronuclei. Chromosoma 2020. [Google Scholar] [CrossRef]
- Mackenzie, K.J.; Carroll, P.; Martin, C.A.; Murina, O.; Fluteau, A.; Simpson, D.J.; Olova, N.; Sutcliffe, H.; Rainger, J.K.; Leitch, A.; et al. cGAS surveillance of micronuclei links genome instability to innate immunity. Nature 2017, 548, 461–465. [Google Scholar] [CrossRef] [Green Version]
- de Oliveira Mann, C.C.; Kranzusch, P.J. cGAS Conducts Micronuclei DNA Surveillance. Trends Cell Biol. 2017, 27, 697–698. [Google Scholar] [CrossRef]
- Spektor, A.; Umbreit, N.T.; Pellman, D. Cell Biology: When Your Own Chromosomes Act like Foreign DNA. Curr. Biol. 2017, 27, R1228–R1231. [Google Scholar] [CrossRef] [PubMed]
- Quek, H.; Luff, J.; Cheung, K.; Kozlov, S.; Gatei, M.; Lee, C.S.; Bellingham, M.C.; Noakes, P.G.; Lim, Y.C.; Barnett, N.L.; et al. A rat model of ataxia-telangiectasia: Evidence for a neurodegenerative phenotype. Hum. Mol. Genet. 2017, 26, 109–123. [Google Scholar] [CrossRef]







Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the author. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Fenech, M. Cytokinesis-Block Micronucleus Cytome Assay Evolution into a More Comprehensive Method to Measure Chromosomal Instability. Genes 2020, 11, 1203. https://doi.org/10.3390/genes11101203
Fenech M. Cytokinesis-Block Micronucleus Cytome Assay Evolution into a More Comprehensive Method to Measure Chromosomal Instability. Genes. 2020; 11(10):1203. https://doi.org/10.3390/genes11101203
Chicago/Turabian StyleFenech, Michael. 2020. "Cytokinesis-Block Micronucleus Cytome Assay Evolution into a More Comprehensive Method to Measure Chromosomal Instability" Genes 11, no. 10: 1203. https://doi.org/10.3390/genes11101203