Abstract
Understanding how populations are structured in space and time is a central question in evolutionary biology. Parasites and their hosts are assumed to evolve together, however, detailed understanding of mechanisms leading to genetic structuring of parasites and their hosts are lacking. As a parasite depends on its host, studying the genetic structure of both parasite and host can reveal important insights into these mechanisms. Here, genetic structure of the monogenean parasite Gyrodactylus thymalli and its host the European grayling (Thymallus thymallus) was investigated in 10 tributaries draining into the large Lake Mjøsa in Norway. The population genetic structure of spawning grayling was studied using microsatellite genotyping, while G. thymalli was studied by sequencing a mitochondrial DNA gene (dehydrogenase subunit 5). Two main genetic clusters were revealed in grayling, one cluster comprising grayling from the largest spawning population, while the remaining tributaries formed the second cluster. For both taxa, some genetic differentiation was observed among tributaries, but there was no clear isolation-by-distance signature. The structuring was stronger for the host than for the parasite. These results imply that moderate to high levels of gene flow occur among the sub-populations of both taxa. The high parasite exchange among tributaries could result from a lack of strong homing behavior in grayling as well as interactions among individual fish outside of the spawning season, leading to frequent mixing of both host and parasite.
Similar content being viewed by others
Introduction
Describing and explaining patterns of genetic population structure continues to be a central theme in evolutionary biology (Richardson et al., 2016). Large genetic variation within and among populations is assumed to be important for adaptive evolutionary change, and in this context gene flow among populations may potentially enhance or hamper local adaptation (Wright, 1943; Slatkin, 1987; Hendry et al., 2000). Understanding the origin and evolution of population genetic structure in natural populations is inherently challenging as variation in e.g. population size, life cycle, generation time and behavior all may be important factors impacting on structure (Nadler, 1995; Barrett et al., 2008).
Most studies of population genetic structure tend to focus on single species, but clearly the structure of many organisms depends on how other species around them are distributed and structured. This should be particularly so for parasites, in particular for those that are highly host specific. For instance, ecto-parasites experience a habitat that may be temporary and varying in quality as the host moves across environments, whereas endo-parasites may experience a more stable environment (Poulin, 2007). Contrasting the genetic structure of the host and its ecto-parasite may illuminate how the genetic structure of the parasite is contingent upon its main host (Huyse et al., 2005; Poulin, 2007; Mazè-Guilmo et al., 2016). However, little is known about the specific mechanisms that facilitate or constrain gene flow in parasite taxa (Poulin, 1997; Blouin et al., 1999). Intuitively, host dispersal is important for facilitating gene flow in parasites (Blouin et al., 1995; McCoy et al., 2012). Parasites with only freshwater hosts (termed an autogenic life cycle) have lower dispersal ability between freshwater localities than parasites transmitted through an alternation between freshwater and terrestrial hosts (allogenic life cycle) (Criscione & Blouin, 2004; Blasco-Costa et al., 2013). How the genetic variation and structure covary in associated hosts and parasites seem to be species-specific and dependent upon type of life cycles (Raeymaekers et al., 2008; Louhi et al., 2010; Song et al., 2014). However, remarkably few studies have compared the genetic structure of fish hosts and their parasites (but see Geist & Kuehn, 2008; Sprehn et al., 2015; Kmentová et al., 2020).
Salmonid fishes are commonly hosts to a variety of ecto-parasites (Pickering & Christie, 1980; Pike & Lewis, 1994). Salmonids often have complex life histories that include ontogenetic habitat shifts and changes in social behavior (Hendry et al., 2004; Hesthagen & Sandlund, 2004). Migration between spawning habitats suitable for egg and juvenile development and feeding habitats suitable for larger fish are common (Hesthagen & Sandlund, 2004; Quinn, 2005; Jonsson & Jonsson, 2011). Natal homing is also typical for salmonids (Stabell, 1984), leading to genetic differentiation of neighboring populations even when there are no physical barriers to dispersal (Fleming & Reynolds, 2004; Waples, 2004). Salmonid fishes often change from being solitary and territorial to moving in groups (schooling). In particular, most salmonids will actively aggregate during spawning (Fleming & Reynolds, 2004).
The European grayling (Thymallus thymallus (L.)), hereafter: grayling, is assumed to be a philopatric species, meaning that it returns to its natal spawning location for reproduction (Kristiansen & Døving, 1996). Such natal homing behavior lead to reduced gene flow among populations and thus genetic structuring (Barson et al., 2009; Koskinen et al., 2001; Junge et al., 2011). During early spring, mature grayling living in lakes aggregate outside the spawning tributaries before they enter the tributary for spawning (Beauchamp, 1990; Northcote, 1995). During the spawning season the males defend territories and during such periods of physical aggregation individuals will be close together and parasite transmission may be more common than during the rest of the life cycle (Pickering & Christie, 1980; Richards et al., 2012). If this is the case, and if homing is strong, one may expect the population genetic structure of host and parasite to be associated. The parasite is here highly contingent upon host homing behavior.
Grayling is the host of the ecto-parasitic monogenean Gyrodactylus thymalli Žitňan, 1960 (Soleng et al., 1999). G. thymalli has an autogenic life cycle where transmission occurs directly between grayling individuals when in close contact (see Bakke et al., 2007). Inter-host contacts are most likely occurring during the spawning time of the grayling (Pickering & Christie, 1980). During the remaining time of the year there will probably be less opportunity for physical contact among grayling individuals facilitating parasite transmission. However, it is documented that Arctic grayling (Thymallus arcticus) in rivers establish dominance hierarchies facilitated by individual interactions (Hughes, 1992). We do not know if such hierarchies are developed for grayling feeding in the littoral zone of lakes. Thus, there is most likely a short spatio-temporal parasite transmission window during the spawning season. If this is the case, it will make the Gyrodactylus thymalli-grayling system a good model system for exploring how genetic structure develops among parasites and hosts.
We have compared the population genetic structure of G. thymalli (using mitochondrial DNA sequencing) and its main host, the grayling (using microsatellite genotyping), in the largest Norwegian lake – Lake Mjøsa. Lake Mjøsa holds numerous tributaries where populations of grayling migrate for spawning during spring (Kristiansen & Døving, 1996). Previous studies have detected significant genetic structuring at large geographic scales for both grayling and G. thymalli (Junge et al., 2014; Pettersen et al., 2015; Van Leeuwen et al., 2018). Here, we investigate the genetic structure at a smaller geographic scale, by contrasting samples of hosts and parasites sampled during the grayling spawning season in ten tributaries to Lake Mjøsa. There are no physical barriers to gene flow among tributaries.
Methods
Study area and sampling
Lake Mjøsa is a large (362 km2) and deep (mean depth 153 m, maximum depth 453 m) lake in southeast Norway (L’Abée-Lund et al., 2009) (Fig. 1). The catchment area is 16,563 km2, containing mountainous areas with glaciers as well as lowland areas heavily influenced by human settlement and agriculture (Haugen et al., 2008). The fish assemblage is relatively rich in a Norwegian context, containing 20 species in total (Hesthagen & Sandlund, 2004). Grayling are common throughout the lake and are known to spawn during spring in a large number of tributaries draining into the lake. A more detailed description of the tributaries is given in Kristiansen & Døving (1996). Grayling in Lake Mjøsa usually become sexually mature at an age of three years and mature grayling migrate to the tributaries for spawning during May and June (Kristiansen & Døving 1996). Grayling usually spawn when the water temperature reaches approximately 5°C (own observations and Fabricius and Gustafson 1955; Parkinson et al., 1999; Gregersen et al., 2008). Generally, the grayling complete spawning within approximately two weeks, and then return to the lake. Grayling embryos hatch in June and the juveniles stay in the tributaries until late summer (Kristiansen & Døving, 1996).
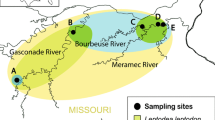
Tributaries in Lake Mjøsa where spawning grayling (Thymallus thymallus) and Gyrodactylus thymalli were sampled. The colour codes refer to relative frequencies of the mtDNA (NADH5) haplotypes of G. thymalli (G08–G31). The tributary codes refer to the names in Table 1. In parentheses are given first the total number of sampled grayling and secondly the number of infected grayling (bold). The inset shows a map of Norway with the location of the Lake Mjøsa indicated by a star.
In total, 146 mature grayling varying in size from 27 to 44 cm were sampled from ten tributaries by electrofishing during the 2008 and 2009 spawning seasons (Fig. 1, Table 1). Five of the tributaries drain into the eastern side of the lake and four into the western side. The last population was sampled in the main inflowing river (Gudbrandsdalslågen), downstream of the Hunderfossen waterfall (label: HUN) in the north. The geographic distances between pairs of tributaries are available in Additional file 1. There are no apparent physical barriers to migration and gene flow among any of the tributaries. A previous mark-recapture study has shown that grayling tend to spawn repeatedly in the same tributary and that they only to a limited degree migrate between the eastern and western side of the lake (Kristiansen & Døving, 1996). We therefore grouped the tributaries into three different units (west, east and north; Table 1) when testing for large-scale geographic population structure. In addition, we tested for differentiation among the individual spawning populations.
All fish were killed quickly by a blow to the head, according to national regulations. All the fins were excised and stored in 96% ethanol for future examination. A small piece of fin tissue from each individual grayling was stored for later DNA-extraction and genotyping. In the lab, the fins of each fish were inspected for the presence of G. thymalli using a stereo-microscope. In total, 129 G. thymalli specimen were sampled for DNA-extraction and sequencing (Table 1). Species of Gyrodactylus are viviparous (Malmberg, 1993) and most or all individuals on one host are likely the offspring of one mother. Thus, we sampled one parasite from each fish, rather than several parasites from the same fish, to maximise sampled genetic variation. A subset (n = 45) of the sampled parasites have been used in a previous study investigating the genetic structure on a much larger geographic scale (Pettersen et al., 2015). All G. thymalli sampled from one tributary are referred to as belonging to one host population.
DNA from fin tissues of the host and whole specimen of the parasite were extracted using the GeneMole® tissue kit (MoleStrips™ DNA) and apparatus (Mole Genetics AS), following the manufacturer’s instructions.
Molecular analyses of G. thymalli
All G. thymalli specimens were sequenced for the mitochondrial dehydrogenase subunit 5 (NADH5) gene. This mtDNA gene has been identified as a highly variable region in G. thymalli (Plaisance et al., 2007; Pettersen et al., 2015). We used the protocol and primer pairs presented in Huyse et al. (2008) to amplify an 894-bp fragment of the NADH5 gene. Polymerase chain reaction (PCR) was performed using PuReTaq ready-to-go PCR beads (GE Healthcare), 1 μmolL−1 of each primer, and 5 μL of the extracted DNA in a 50-μL reaction volume. PCR products were purified using 10 × diluted exoSAP-IT (USB). Cycle sequencing, using the same primers as in the PCR reaction, was performed in 10-μL reactions using 2-μLBigDye terminator cycle sequencing ready reaction kit (Applied Biosystems), 2 μL 5 × sequencing buffer, 10 pmol primer, and 3-μL cleaned PCR product. The cycle sequencing was done on an ABI 3730 high-throughput capillary electrophoresis machine. All sequences were edited using Sequencher 5.0 and aligned with the ClustalW algorithm in MEGA 5.0 (Tamura et al., 2007), with default parameter settings. Genetic distances were calculated according to the Tamura (1992) three-parameter + Gamma (T92 + G) model. This model gave the best fit in a model selection test of 24 different nucleotide substitution models, selected with models with lowest BIC scores (Bayesian information criterion) in MEGA 5.0 (Tamura et al., 2007; Tamura et al., 2004). For each codon, we estimated the number of synonymous and non-synonymous substitutions produced using maximum likelihood (ML) analysis of natural selection codon-by-codon using HyPhy in MEGA 5.0 (Felsenstein, 1981; Muse & Gaut, 1994; Pond & Frost, 2005; Suzuki & Gojobori, 1999). Further, Tajima`s neutrality test was performed on each population in MEGA 5.0 to test if sequences deviated from neutrality expectations (i.e. equilibrium between mutation and genetic drift; (Tajima, 1989). Deviations from neutrality are indicated by p-values below 0.05 in MEGA 5.0.
An analysis of molecular variance (AMOVA) for haplotypes was conducted in GenAlEx 6.2 (Excoffier et al., 1992; Peakall & Smouse, 2006; Peakall & Smouse, 2012) partitioning the variance components among tributaries, among individuals within each tributary, and between tributaries on the eastern and western side of Lake Mjøsa. Additionally, GenAlEx was used for an analysis of pairwise genetic differentiation (population haploid pairwise genetic differentiation φST analog to FST) between tributaries. The φST was then used in a principal component analysis (PCA) to visualize potential G. thymalli groupings (Peakall & Smouse, 2006).
In order to test if there were significant differences in the number of sampled haplotypes among the three regions (east, north and west) a sample-based rarefaction analysis in Estimate S (Version 8.2.0) was used correcting for sample size. To infer the relationships among haplotypes a median-joining network was estimated in NETWORK 4.5.1.6 (Flexus Technology). MEGA 5.0 was used to test for the phylogenetic relationships among haplotypes using Neighbor-Joining (NJ) (Saitou & Nei, 1987) and ML with a bootstrap consensus tree inferred from 1,000 replicates (Felsenstein, 1985) using the same molecular evolution model as before (Tamura three-parameter + Gamma (T92 + G)). Two sequences were used as outgroups; G. thymalli from the type locality (River Hnilec, Slovakia; GenBank number EF527269) and a more phylogenetically distant species G. derjavinoides Malmberg, Collins et al., 2007 (GenBank number EU293891) sampled from brown trout (Salmo trutta L.).
Molecular analyses of grayling
All grayling individuals were genotyped for a set of 17 microsatellite loci following Junge et al. (2010). Methodological details and marker information are found in Additional file 2. Briefly, we ran eight PCRs (using 1 × Qiagen Multiplex PCR Master Mix) with annealing temperatures between 58°C and 60°C. Subsequently, PCRs were combined in two running sets, with three and five PCRs, for electrophoresis on an ABI3730xl Genetic Analyzer, Applied Biosystems (ABI, California, USA). Genotypes were scored using GeneMapper 4.0 software (ABI).
We tested for neutrality of the 17 grayling microsatellites using Lositan (Antao et al., 2008; Beaumont & Nichols, 1996). GenAlEx 6.41(Peakall & Smouse, 2012) was used to estimate grayling microsatellite diversity (frequencies and mean of alleles per locus, observed and expected heterozygosity). To calculate allelic richness the program FSTAT 2.9.3.2 (Goudet, 1995) was used, with the smallest population size set at 8 individuals. Deviation from Hardy–Weinberg and linkage equilibrium was tested using GENEPOP version 4.0.7 (Rousset, 2008). Markov Chain (MC) parameters chosen for the test were: 10,000 dememorization number and maximum number of iterations per batch for 1,000 batches. To avoid type II errors we applied sequential Bonferroni corrections (Moran, 2003).
To determine the genetic structure, a number of different approaches were used. First, an AMOVA was performed using the locus-by-locus procedure in Arlequin 3.5.1.3 (Excoffier et al., 1992; Excoffier et al., 2005) to test for genetic variation across tributaries and/or regions (west/east). Here, the variance components were partitioned into (i) among regions (west/east), (ii) among tributaries and (iii) among individuals within tributaries. The analysis was conducted for the complete dataset, but excluding the northern population HUN, as it was a priori expected to be genetically distinct from the others. The program POWSIM was used to assess the statistical power, by mimicking sampling from populations at a predefined level of expected divergence through random number computer simulations under a classical Wright-Fisher model without migration or mutation. Significance of estimates was based on 1,000 independent simulations.
The program GENEPOP (Weir & Cockerham, 1984) was used to test for genetic differentiation among each population pair at every locus and over all loci by an exact G tests. As described above we used the same MC parameter settings and multiple test correction procedures. We also used GENEPOP to estimate pairwise and global FST (Weir & Cockerham, 1984).
The program STRUCTURE 2.3 (Pritchard et al., 2000) was applied to infer population structuring. Ten replicate runs for K values between 1 and 9, using the admixture model, the LOCPRIOR option (Hubisz et al., 2009) and assuming correlated allele frequencies (Pritchard et al., 2000; Falush et al., 2003) were performed with a burnin of 500,000 and 1,000,000 MCMC iterations. The analyses were performed both for the complete dataset and for the dataset with the HUN sampling location excluded. HUN clearly grouped distinctly from the other populations.
Grayling and G. thymalli
We tested for an isolation-by-distance (IBD) genetic structure (Wright, 1943) in both grayling and G. thymalli using a Mantel test implemented in GenAlEx 6.41 (Peakall & Smouse, 2006; Peakall & Smouse, 2012) by correlating genetic distances (FST/(1 − FST)) or (φST/(1 − φST)) (Rousset, 1997) with geographic distances (km). Two distance metrics were tested, one measure of geographic distances was the shortest water distance between tributary mouths and second measure was the shortest distance along the shoreline (see Additional file 1). These tests were conducted for both grayling and G. thymalli separately. Shoreline distances were used in addition to the shortest distance since mark-recapture studies in grayling in Lake Mjøsa have shown that grayling usually disperse along shorelines rather than across open water (Kristiansen & Døving, 1996). The level of significance was evaluated by performing 9,999 permutations.
In addition, we tested if the pairwise genetic distances between hosts (FST/(1 − FST)) and between parasites (φST/(1 − φST)) were correlated using a Mantel test in GenAlEx 6.41.
Results
G. thymalli
A total of 129 individual G. thymalli were sampled from the grayling spawning in the 10 tributaries to Lake Mjøsa (Table 1). The mtDNA alignment of the NADH5 gene from G. thymalli (768 bp, GenBank no. KY264209-KY264338, Additional file 3) revealed 13 haplotypes, defined by 16 polymorphic sites, leading to 9 amino acid substitutions (Table 1). All the haplotypes have been detected previously in a large-scale study encompassing the total River Glomma watershed (Pettersen et al., 2015). The nucleotide frequencies on NADH5 were 31% (A), 31% (T/U), 16% (C), and 22% (G). The polymorphic sites of NADH5 showed no indication of selection on amino acid triplets based on the codon-by-codon ML analysis.
Sample size, allele frequency (Af), number of segregating sites (S), nucleotide diversity (π), and Tajima`s D test statistics from the different tributaries can be found in Table 1. None of the populations showed divergence from neutrality when tested by Tajima`s D. The two most common haplotypes (G8 and G24) were found in almost all tributaries (Fig. 1). Haplotype diversity was slightly higher on the western side of Lake Mjøsa (8) compared to the eastern side (5). However, the sample-based rarefaction analysis showed overlap in confidence intervals between the number of haplotypes in the western (mean [95% CI]; 10 [6.7–13.3]), eastern (5 [1.7–8.7]) and northern HUN (5 [1.8–8.5]) populations. Private alleles were observed in three different populations; SKU, STA and HUN.
The AMOVA revealed that 94% (VC: 1.233, φST = 0.052, P = 0.030) of the haplotype variance could be explained by within-tributary variation, whereas only 0% (VC: 0.00, φCT = − 0.010, p = 0.680) of the variation could be explained by region (West/East) and 6% (VC: 0.080, φSC = 0.061, P = 0.020) by among-tributary variation.
The network analysis on NADH5 resulted in a star-shaped network with no signal of phylogeographic structuring (Fig. 2). The network analysis detected one unsampled ancestor node (mv1) with four branches with the two most common haplotypes (G24 and G8) in different branches. There was no support for phylogenetic relationships between G. thymalli haplotypes within Lake Mjøsa based on the NJ method and ML method, evidenced by low bootstrap support (i.e. bootstrap estimates below 70%, (Hillis & Bull, 1993).
Median-joining network for the NADH5 haplotypes of Gyrodactylus thymalli. Haplotypes colour-coded as in Fig. 1. The cross lines indicate the mutation steps and the size of the circles represents haplotype frequency. The red node mv1- is the hypothetical unsampled ancestor haplotype
We found weak evidence for some pairwise genetic differentiation among G. thymalli sampled in the different tributaries; i.e. 5 out of 45 estimates were significant (Table 2). φST values varied between 0.109–0.764 which is significant given the Bernoulli method. The sample from the GAU tributary was involved in three of these five significant comparisons. There was no relationship between pairwise genetic differentiation (measured as φST/(1 − φST) and pairwise geographic distance measured as either the shortest distance across water (R2 = 0.015, pMantel = 0.636) or the shortest distance along the shoreline (R2 = 0.008, pMantel = 0.220). This means there was no signal of IBD.
A PCA on the population genetic distances revealed that the populations could not be grouped by habitat/geography (Additional file 4).
Grayling
All the microsatellite loci confirmed to neutral expectations when tested and were therefore included in the analyses (Additional file 5). The average number of alleles per locus within a sampling site varied between 4.6 and 6.4 (Additional file 6). Allelic richness (AR) was calculated for all tributaries and ranged from 4.2 to 5.0 (Table 1). Observed (Ho) and expected (He) heterozygosity ranged from 0.56 to 0.64, and 0.60 to 0.67, respectively (Table 1). All populations confirmed to Hardy–Weinberg equilibrium and all locus pairs showed linkage disequilibrium, i.e. they were not linked.
Out of 45 pairwise population comparisons for genetic differentiation, a total of 29 (64%) comparisons showed significant differentiation (Table 2), which is significant given the Bernoulli method (i.e. the probability of getting this result with α = 0.05 by chance alone is very small). The HUN population was significantly differentiated from all the other nine populations. For the remaining samples from Lake Mjøsa tributaries, BAL was significantly differentiated from all other samples except STA (its closest neighboring tributary), whereas FJA, HAM and VIK were differentiated from all but two other tributaries (RIN and STA).
The AMOVA revealed that close to 99% (VC: 5.412, FCT = 0.001, P < 0.5) of the genotype variation could be assigned to within-sample variation, whereas 0% (variance component, VC: 0.058, FST = 0.012, P < 0.01) could be explained by region (west/east) and 1% (VC: 0.064, FSC = 0.012, P < 0.01) by among tributary variation. The global FST (jackknifed over loci) was 0.033 (95% CI 0.004–0.020, 99% CI 0.002–0.024), and pairwise FST-values varied between − 0.008 and 0.094 (Table 2).
There was no clear isolation-by-distance signal, but a weak trend was observed when using the complete dataset and shortest waterway distance as the spatial metric (R2 = 0.530, pMantel = 0.056, Fig. 3; see Additional file 7 for shortest shoreline distances, R2 = 0.014, pMantel = 0.126). After excluding the HUN population (assumed to be a resident river-living population), the relationship between genetic and geographic distance remained non-significant (waterway distance: R2 = 0.003, pMantel = 0.397, shoreline distance: R2 = 0.003, pMantel = 0.335).
The STRUCTURE analysis revealed that the most likely number of genetic clusters were K = 2 (Additional file 8), separating the large tributary HUN from the nine smaller tributaries which jointly composed the second genetic cluster (Additional file 9, 2A). Removing the HUN population from the data set and rerunning the STRUCTURE analysis revealed no additional sub-structure within the nine tributaries (Additional file 9, 2B).
Host and parasite comparison
There was no correlation between the pairwise genetic distances of G. thymalli and that of its grayling host (R2 = 0.010, pMantel = 0.470).
Discussion
This study shows a lack of strong genetic structure within both G. thymalli and its host the grayling when comparing samples from ten tributaries to the large Lake Mjøsa in Norway. For grayling, two genetic clusters were evident, one comprising fish from the largest inlet tributary while fish from the remaining nine tributaries formed the second cluster. Weak but significant genetic differentiation among the majority of tributaries was observed for grayling, whereas there was no evidence for structuring in G. thymalli. Further, there was no correlation between the pairwise genetic distances of the hosts and those of the parasites, which might indicate that different mechanisms determine their level of gene flow and thus population structure.
Genetic structure of G. thymalli
A total of 13 mtDNA haplotypes were found in G. thymalli in Lake Mjøsa. Most of these haplotypes were shared among parasites found in the ten grayling spawning populations, except for a few endemic haplotypes. Further, there was no clear geographic structure, neither when tested among the three geographic groups (east–west-north) nor among tributaries. All the haplotypes have been observed earlier in a large-scale study of G. thymalli in the River Glomma watercourse (Pettersen et al., 2015). Lake Mjøsa is part of this river system. The genetic network that was found in Lake Mjøsa is very similar to the network described for the total River Glomma system (Pettersen et al., 2015). Thus, none of the haplotypes in G. thymalli have likely arisen post-glacially in Lake Mjøsa.
There was low support for a geographic structuring of G. thymalli haplotypes among tributaries to Lake Mjøsa based on the Neighbor-Joining method. This was also supported by the EstimateS analysis comparing haplotype diversity adjusting for sample size, and by the AMOVA analysis showing that most of the genetic variation was due to the within-tributary effects. The PCA plot of genetic distances also supported a lack of differentiation. These results all imply that there is a low degree of genetic differentiation among G. thymalli sub-populations in Lake Mjøsa.
No similar studies of genetic variation of G. thymalli in lake ecosystems can be found in the literature. However, a study of 309 G. thymalli from 20 sites in the large Glomma river system found 35 distinct NADH5-haplotypes (Pettersen et al., 2015). Further, Lindqvist et al. (2007) observed three G. thymalli NADH5-haplotypes on a sample of 16 grayling in one site in a tributary. In contrast to the finding of several haplotypes of G. thymalli within different tributaries in our study, only one single haplotype of G. salaris and G. thymalli was found on several populations of Atlantic salmon (Salmo salar L.) and grayling in northern Baltic rivers (Kuusela et al., 2009). Kuusela et al. (2009) related their findings to the fugitive strategy hypothesis (Platt & Weis 1985). This hypothesis suggests that when a river is already occupied by a parasite strain (or haplotype), there is reduced probability for other strains to invade (Kuusela et al., 2009). This situation is related to the concepts of invasion dynamics, the probability of successfully invading an already occupied habitat, and the waiting time until dominance of the invader (Gavrilets & Gavrilets, 2004). There is no support for the fugitive strategy hypothesis in our extensive study. A potential explanation for a one-tributary-one-haplotype observation can be low sampling effort (Walther et al., 1995), as increased sample size leads to an increased number of haplotypes observed (Gavrilets & Gavrilets, 2004).
Genetic structure of grayling
The results suggest that two well-defined genetic clusters of grayling are present in Lake Mjøsa. Grayling sampled in the largest tributary to Lake Mjøsa (HUN) were clearly differentiated from grayling spawning in the nine other tributaries. This finding was also supported by the observed isolation-by-distance trend where the genetic distinctiveness of the HUN tributary was evident. Grayling in the HUN tributary could belong to a stationary river-living population, in contrast to tributary-spawning but lake-living grayling in the other tributaries. This could reduce the probability for gene flow between grayling populations belonging to these two clusters. Furthermore, the HUN tributary is by far the largest in terms of water flow and probably population size, meaning that gene flow from other tributaries likely would have minimal effect on the population structure of grayling in the HUN tributary. However, some gene flow from HUN to other tributaries is expected as indicated in the STRUCTURE plot (see Additional file 7). Junge et al. (2011) studied the population structure of grayling in Lake Lesjaskogsvatnet, located upstream the locality HUN. By studying genetic differentiation among these populations over several years they observed significant among-tributary genetic differentiation even if the level of differentiation was low. This is similar to our observations in Lake Mjøsa. Thus, the grayling in the two lakes are similar in partitioning most genetic variation to within tributaries. Further, there was a weak isolation-by-distance signal when all the populations were included in the analysis. However, the signature became non-significant when the HUN-population was excluded. In comparison, Junge et al. (2011) detected a significant isolation-by-distance pattern for grayling spawning in different tributaries to the much smaller Lake Lesjaskogsvatnet. This could be due to the large environmental differences between tributaries in Lake Lesjaskogsvatnet, potentially leading to stronger selection for natal homing.
The low genetic differentiation seen among the Lake Mjøsa grayling was a little surprising as an earlier mark-recapture study documented moderate to strong homing behavior of grayling in Lake Mjøsa (Kristiansen & Døving, 1996). Further, that study also found that grayling mainly used the littoral zone nearby the tributary where they were initially marked when feeding during summer (Kristiansen & Døving, 1996). The same study found that straying among spawning tributaries was 16% (an overall estimate), and only 2% migration between the western and eastern side of the lake. A recent telemetry study in Lake Lesjaskogvatn has documented a relatively small home range for lake-living grayling (mean 0.58 km2) (Bass et al., 2014). Further, a study of the feeding and habitat use showed that grayling mainly used the littoral zone (Haugen & Rygg, 1996). Thus, given the apparent littoral zone preference of grayling and the moderate to strong homing behavior to spawning tributaries in Lake Lesjasjkogsvatnet, we expected a higher degree of genetic differentiation in the Lake Mjøsa grayling.
Genetic studies have also been conducted on brown trout spawning in several tributaries to Lake Mjøsa (Skaala, 1992; Linløkken et al., 2014). Brown trout from the different spawning tributaries were significantly differentiated, with clear difference between east and west populations (Skaala, 1992). Further, there seemed to be a significant isolation-by-distance pattern (Linløkken et al., 2014). Thus, brown trout seems more genetically differentiated than grayling in spawning tributaries to Lake Mjøsa. Brown trout spawn during autumn, with incubation of eggs and embryos during winter and spring. The importance of being locally adapted may thus be stronger, leading to stronger selection for natal homing than in a spring-spawning fish having rapid development during spring and early summer. Also, for grayling spawning in large rivers, low levels of natal homing and genetic structuring have been found, opposite to what is common for brown trout (Junge et al., 2014; Van Leeuwen et al., 2018).
Grayling—G. thymalli comparisons
There was no correlation between the pairwise genetic distances of grayling and G. thymalli sampled in the different spawning tributaries to Lake Mjøsa. Based on the finding that grayling repeatedly spawn in the same tributaries year after year (Kristiansen & Døving, 1996), we expected that grayling populations should be genetically differentiated and that G. thymalli should follow that same pattern. The estimates of pairwise genetic differentiation among spawning tributaries were significant in 64% of the cases in grayling (FST) and in only 11% in G. thymalli (φST). This pattern resembles the pattern found by Song et al. (2014) who compared an acanthocephalan (Acanthosentis cheni Amin, 2005) parasite and its fish host the Japanese grenadier anchovy (Coili nasus Temminck & Schlegel, 1846). They observed a higher level of gene flow among the parasites than among hosts. This may be a general phenomenon in host-parasite systems (Anton et al., 2007; Levin & Parker, 2013; McCoy et al., 2012). However, in contrast, Criscione & Blouin, (Criscione & Blouin, 2007) studied the trematode parasite (Plagioporus shawi (McIntosh, 1939) Margolis, 1972) and its salmonid hosts (Oncorhynchus spp.) and observed congruent genetic structure and divergence between the host and parasite.
Nieberding et al. (2004) suggested that the genetic variation of a parasite can be used to provide more in-depth information on the phylogeographic history of its host. They suggested that the “magnifying glass” hypothesis, suggesting higher genetic variation and differentiation in the parasite compared to the host, could render the study of the genetic structure of parasites useful for investigating the structure of its host (Mackenzie, 1987; MacKenzie, 2002). In our system, however, the host showed lower levels of gene flow among populations than did the parasite. This led to some genetic structuring in grayling, but less in G. thymalli. These results do not support the “magnifying glass” hypothesis, but are similar the findings in another Gyrodactylus - host system (Huyse et al., 2017). That study compared the genetic variation of G. gondae Huyse, Malmberg & Volckaert 2005 and its sand goby host Pomatoschistus minutus (Pallas, 1770) along the Atlantic coasts of Europe, a much larger area than our study system (Huyse et al., 2017). The main findings were that the parasite populations were significantly differentiated, but the structuring was not consistently higher than in its host populations (Huyse et al., 2017). A recent study of monogeneans (Dactylogyridae) parasitize clupeid fishes in Lake Tanganyika also did not find support for the “magnifying glass” hypothesis (Kmentová et al., 2020). Rather, the parasites did not show any clear geographical population structure. Overall, there seems to be little support for the magnifying glass hypothesis.
It is commonly suggested that the genetic structure of parasites is congruent with that of their hosts, because the host dispersal limits parasites dispersion pattern (e.g. Jarne & Theron, 2001, review by Mazè-Guilmo et al., 2016). However, a meta-study covering diverse taxonomic groups of hosts and parasites, concluded that the genetic differentiation on average was lower in parasites than their hosts (Mazè-Guilmo et al., 2016). This result corresponds well with findings in the present study. Genetic differentiation depends on generation time and population size. G. thymalli has a shorter generation time than grayling (Bakke et al. 2007), and thus stronger genetic differentiation may be expected. However, genetic structure is weaker in parasites with asexual reproduction compared to parasites with sexual reproduction (Mazè-Guilmo et al., 2016). G. thymalli basically reproduce asexually, thus leading to lower overall differentiation rates (Fox et al., 1996; Huyse et al., 2005; Liu et al., 1996).
We assumed that the between-host transmission of G. thymalli mainly occurs when grayling is spawning. In our study we thus sampled grayling only during the spawning season, when grayling concentrate on the spawning areas and interact intensively. However, transmission of parasites among hosts can also occur during time periods outside the spawning season, as seen in grayling when they search for food in the littoral area (Eloranta et al., 2011). In a study by Richards et al. (2012) transmission of Gyrodactylus spp. on guppies were dependent upon the social behavior of the fish where the most parasitized fish had the highest shoaling intensity. Grayling may develop dominance hierarchies, at least in rivers (Hughes, 1992). Such social interactions may lead to parasite transmission. Further, if grayling individuals aggregate while feeding on abundant food resources in the littoral zone transmission may also occur. Thus, transmission of G. thymalli can likely occur also outside of the spawning season, facilitating a more effective transmission of the parasite. Studies on the social behavior of grayling outside of the spawning season is needed.
Conclusions
A moderate to high level of gene flow exists among spawning populations of grayling and among the populations of its ectoparasite G. thymalli. This led to some genetic structuring in grayling, but less so in G. thymalli. This is contrary to our expectation of a moderate to strong association between host-parasite genetic structures and genetic distances. The low genetic divergence in G. thymalli could be due to weak homing behavior in grayling or more social interaction between individuals also outside of the spawning season. The exact mechanisms for the transmission of G. thymalli between grayling host individuals will need to be investigated in the future, taking space and time into consideration.
Data availability
All relevant data are either available in the Additional files 5 and 6 (microsatellite data for grayling) or Additional file 3 (mtDNA haplotypes for Gyrodactylus thymalli). Sequence data are also downloaded at Genbank; accession numbers KY264209-KY264338.
References
Antao, T., A. Lopes, R. J. Lopes, A. Beja-Pereira & G. Luikart, 2008. LOSITAN: a workbench to detect molecular adaptation based on a Fst-outlier method. BMC Bioinformatics 9: 5.
Anton, C., I. Zeisset, M. Musche, W. Durka, J. J. Boomsma & J. Settele, 2007. Population structure of a large blue butterfly and its specialist parasitoid in a fragmented landscape. Molecular Ecology 16(18): 3828–3838.
Bakke, T. A., J. Cable & P. D. Harris, 2007. The biology of gyrodactylid monogeneans: the “Russian-doll killers”. Advances in Parasitology 64: 161–330.
Barrett, L. G., P. H. Thrall, J. J. Burdon & C. C. Linde, 2008. Life history determines genetic structure and evolutionary potential of host-parasite interactions. Trends in Ecology & Evolution 23(12): 678–685.
Barson, N. J., T. O. Haugen, L. A. Vøllestad & C. R. Primmer, 2009. Contemporary isolation by ditance, but not isolation by time, among demes of European grayling (Thymallus Thymallus, Linneus) with recent common ancestors. Evolution 63(2): 549–556.
Bass, A. L., T. O. Haugen & L. A. Vøllestad, 2014. Distribution and movement of European grayling in a subarctic lake revealed by acoustic telemetry. Ecology of Freshwater Fish 23(2): 149–160.
Beauchamp, D. A., 1990. Movements, habitat use, and spawning of arctic grayling in a Subalpin Lake Tributary. Northwest Science 64(4): 195–207.
Beaumont, M. A. & R. A. Nichols, 1996. Evaluating loci for use in the genetic analysis of population structure. Proceedings of the Royal Society B: Biological Sciences 263(1377): 1619–1626.
Blasco-Costa, I., A. V. Koehler, A. Martin & R. Poulin, 2013. Upstream-downstream gradient in infection levels by fish parasites: a common river pattern? Parasitology 140(2): 266–274.
Blouin, M. S., C. A. Yowell, C. H. Courtney & J. B. Dame, 1995. Host Movment and the Genetic Structure of Populations of Parasitic Nematodes. Genetics 141(3): 1007–1014.
Blouin, M. S., J. Liu & R. E. Berry, 1999. Life cycle variation and the genetic structure of nematode populations. Heredity 83: 253–259.
Criscione, C. D. & M. S. Blouin, 2004. Life cycles shape parasite evolution: comparative population genetics of salmon trematodes. Evolution 58(1): 198–202.
Criscione, C. D. & M. S. Blouin, 2007. Parasite phylogeographical congruence with salmon host evolutionarily significant units: implications for salmon conservation. Molecular Ecology 16(5): 993–1005.
Eloranta, A. P., A. Siwertsson, R. Knudsen & P.-A. Amundsen, 2011. Dietary plasticity of Arctic charr (Salvelinus alpinus) facilitates coexistence with competitively superior European whitefish (Coregonus lavaretus). Ecology of Freshwater Fish 20(4): 558–568.
Excoffier, L., P. E. Smouse & J. M. Quattro, 1992. Analyse of molecular variance innferred from metric distances among DNA haplotypes: application to human mitochondrial DNA restriction data. Genetics 131(2): 479–491.
Excoffier, L., A. Estoup & J. M. Cornuet, 2005. Bayesian analysis of an admixture model with mutations and arbitrarily linked markers. Genetics 169(3): 1727–1738.
Fabricius, E. & K. J. Gustafson, 1955. Observations on the spawning behaviour of the grayling, Thymallus thymallus (L.). Report Institute of Freshwater Research Drottningholm No. 36: 75–103.
Falush, D., M. Stephens & J. K. Pritchard, 2003. Inference of population structure using multilocus genotype data: linked loci and correlated allele frequencies. Genetics 164(4): 1567–1587.
Felsenstein, J., 1981. Evolutionary trees from gene frequencies and quantitative characters: finding maximum likelihood estimates. Evolution 35(6): 1229–1242.
Felsenstein, J., 1985. Confidence limits on phylogenies: an approach using the bootstrap. Evolution 39(4): 783–791.
Fleming, I. A. & J. D. Reynolds, 2004. Salmonid breeding systems. Oxford University Press, Oxford.
Fox, J. A., M. F. Dybdahl, J. Jokela & C. M. Lively, 1996. Genetic structure of coexisting sexual and clonal subpopulations in a freshwater snail (Potamopyrgus antipodarum). Evolution 50(4): 1541–1548.
Gavrilets, S. & S. Gavrilets, 2004. Fitness landscapes and the origin of species. Princeton, Princeton University Press.
Geist, J. & R. Kuehn, 2008. Host-parasite interactions in oligotrophic stream ecosystems: the roles of life-history strategy and ecological niche. Molecular Ecology 17(4): 997–1008.
Goudet, J., 1995. FSTAT (Version 1.2): a computer program to calculate F-statistics. Journal of Heredity 86(6): 485–486.
Gregersen, F., T. O. Haugen & L. A. Vøllestad, 2008. Contemporary egg size divergence among sympatric grayling demes with common ancestors. Ecology of Freshwater Fish 17(1): 110–118.
Haugen, T. O. & T. A. Rygg, 1996. Food- and habitat-segregation in sympatric grayling and brown trout. Journal of Fish Biology 49(2): 301–318.
Haugen, T. O., P. Aass, N. C. Stenseth & L. A. Vøllestad, 2008. Changes in selection and evolutionary responses in migratory brown trout following the construction of a fish ladder. Evolutionary Applications 1(2): 319–335.
Hendry, A. P., J. K. Wenburg, P. Bentzen, E. C. Volk & T. P. Quinn, 2000. Rapid evolution of reproductive isolation in the wild: evidence from introduced salmon. Science 290(5491): 516–518.
Hendry, A. P., V. Castric, M. T. Kinnison & T. P. Quinn, 2004. The evolution of philopatry and dispersal: homing versus straying in salmonids. Oxford University Press, Oxford.
Hesthagen, T. & O. T. Sandlund, 2004. Fish distribution in a mountain area in south-eastern Norway: human introductions overrule natural immigration. Hydrobiologia 521(1–3): 49–59.
Hillis, D. M. & J. J. Bull, 1993. An empirical-test of bootstrapping as a method for assessing confidence in phylogenetic analysis. Systematic Biology 42(2): 182–192.
Hubisz, M. J., D. Falush, M. Stephens & J. K. Pritchard, 2009. Inferring weak population structure with the assistance of sample group information. Molecular Ecology Resources 9(5): 1322–1332.
Hughes, N. F., 1992. Ranking of feeding positions by drift-feeding Arctic grayling (Thymallus arcticus) in dominance hierarchies. Canadian Journal of Fisheries and Aquatic Sciences 49(10): 1994–1998.
Huyse, T., R. Poulin & A. Theron, 2005. Speciation in parasites: a population genetics approach. Trends in Parasitology 21(10): 469–475.
Huyse, T., K. Buchmann & D. T. J. Littlewood, 2008. The mitochondrial genome of Gyrodactylus derjavinoides (Platyhelminthes: monogenea) – A mitogenomic approach for Gyrodactylus species and strain identification. Gene 417(1–2): 27–34.
Huyse, T., M. Oeyen, M. H. D. Larmuseau & F. A. M. Volckaert, 2017. Co-phylogeographic study of the flatworm Gyrodactylus gondae and its goby host Pomatoschistus minutus. Parasitology International 66(2): 119–125.
Jarne, P. & A. Theron, 2001. Genetic structure in natural populations of flukes and snails: a practical approach and review. Parasitology 123: S27–S40.
Jonsson, B. & N. Jonsson, 2011. Ecology of Atlantic salmon and brown trout: habitat as a template for life histories. Fish and Fisheries Series 33: 1–655.
Junge, C., C. R. Primmer, L. A. Vøllestad & E. H. Leder, 2010. Isolation and characterization of 19 new microsatellites for European grayling, Thymallus thymallus (Linnaeus, 1758), and their cross-amplification in four other salmonid species. Conservation Genetics Resources 2: 219–223.
Junge, C., L. A. Vøllestad, N. J. Barson, T. O. Haugen, J. Otero, G. P. Saetre, E. H. Leder & C. R. Primmer, 2011. Strong gene flow and lack of stable population structure in the face of rapid adaptation to local temperature in a spring-spawning salmonid, the European grayling (Thymallus thymallus). Heredity 106(3): 460–471.
Junge, C., J. Museth, K. Hindar, M. Kraabøl & L. A. Vøllestad, 2014. Assessing the consequences of habitat fragmentation for two migratory salmonid fishes. Aquat Conserv. https://doi.org/10.1002/aqc.2391.
Kmentová, N., S. Koblmüller, M. Van Steenberge, J. A. M. Raeymaekers, T. Artois, E. L. R. De Keyzer, L. Milec, F. Muterezi Bukinga, T. Mulimbwa N’sibula, P. Masilya Mulungula, G. Ntakimazi, F. A. M. Volckaert, M. Gelnar & M. P. M. Vanhove, 2020. Weak population structure and recent demographic expansion of the monogenean parasite Kapentagyrus spp. infecting clupeid fishes of Lake Tanganyika, East Africa. The International Journal for Parasitology 50(6): 471–486.
Koskinen, M. T., J. Piironen & C. R. Primmer, 2001. Interpopulation genetic divergence in European grayling (Thymallus thymallus, Salmonidae) at a microgeographic scale: implications for conservation. Conservation Genetics 2(2): 133–143.
Kristiansen, H. & K. B. Døving, 1996. The migration of spawning stocks of grayling Thymallus thymallus, in Lake Mjøsa, Norway. Environmental Biology of Fishes 47(1): 43–50.
Kuusela, J., R. Holopainen, M. Meinilä, P. Anttila, P. Koski, M. S. Ziętara, A. Veselov, C. R. Primmer & J. Lumme, 2009. Clonal structure of salmon parasite Gyrodactylus salaris on a coevolutionary gradient on Fennoscandian salmon (Salmo salar). Annales Zoologici Fennici 46(1): 21–33.
L’Abée-Lund, J. H., J. A. Eie, P. E. Faugli, S. Haugland, N. A. Hvidsten, A. Jensen, K. Melvold, V. Pettersen, L. E. Petterson & S. J. Saltveit, 2009. Rivers of Boreal Uplands. Elsevier, Amsterdam.
Levin, I. I. & P. G. Parker, 2013. Comparative host-parasite population genetic structures: obligate fly ectoparasites on Galapagos seabirds. Parasitology 140(9): 1061–1069.
Lindqvist, C., L. Plaisance, T. A. Bakke & L. Bachmann, 2007. Mitochondrial DNA variation of a natural population of Gyrodactylus thymalli (Monogenea) from the type locality River Hnilec, Slovakia. Parasitology Research 101(5): 1439–1442.
Linløkken, A. N., W. Johansen & R. Wilson, 2014. Genetic structure of brown trout, Salmo trutta, populations from differently sized tributaries of Lake Mjøsa in south-east Norway. Fisheries Management and Ecology 21: 10.
Liu, Y. C., P. Cortesi, M. L. Double, W. L. MacDonald & M. G. Milgroom, 1996. Diversity and multilocus genetic structure in populations of Cryphonectria parasitica. Phytopathology 86(12): 1344–1351.
Louhi, K. R., A. Karvonen, C. Rellstab & J. Jokela, 2010. Is the population genetic structure of complex life cycle parasites determined by the geographic range of the most motile host? Infection, Genetics and Evolution 10(8): 1271–1277.
Mackenzie, K., 1987. Parasites as indicators of host populations. The International Journal for Parasitology 17(2): 345–352.
MacKenzie, K., 2002. Parasites as biological tags in population studies of marine organisms: an update. Parasitology 124: S153–S163.
Malmberg, G., 1993. Gyrodactylidae and Gyrodactylosis of Salmonidae. Bulletin français de la pêche et de la pisciculture 328: 5–46.
Mazè-Guilmo, E., S. Blanchet, K. D. McCoy & G. Loot, 2016. Host dispersal as the driver of parasite genetic structure: a paradigm lost? Ecology Letters 19(3): 336–347.
McCoy, K. D., P. Beis, A. Barbosa, J. J. Cuervo, W. R. Fraser, J. Gonzalez-Solis, E. Jourdain, M. Poisbleau, P. Quillfeldt, E. Leger & M. Dietrich, 2012. Population genetic structure and colonisation of the western Antarctic Peninsula by the seabird tick Ixodes uriae. Marine Ecology Progress Series 459: 109–120.
Moran, M. D., 2003. Arguments for rejecting the sequential Bonferroni in ecological studies. Oikos 100(2): 403–405.
Muse, S. V. & B. S. Gaut, 1994. A likelihood approach for comparing synonymous and nonsynonymous nucleotide substitution rates, with application to the chloroplast genome. Molecular Biology and Evolution 11(5): 715–724.
Nadler, S. A., 1995. Microevolution and the genetic-structure of parasie populations. Journal of Parasitology 81(3): 395–403.
Nieberding, C., S. Morand, R. Libois & J. R. Michaux, 2004. A parasite reveals cryptic phylogeographic history of its host. Proceedings of the Royal Society B: Biological Sciences 271(1557): 2559–2568.
Northcote, T. G., 1995. Comparative biology and management of Arctic and European grayling (Salmonidae, Thymallus). Reviews in Fish Biology and Fisheries 5(2): 141–194.
Parkinson, D., J. C. Philippart & E. Baras, 1999. A preliminary investigation of spawning migrations of grayling in a small stream as determined by radio-tracking. Journal of Fish Biology 55(1): 172–182.
Peakall, R. & P. E. Smouse, 2006. GENALEX 6: genetic analysis in Excel. Population genetic software for teaching and research. Molecular Ecology Notes 6(1): 288–295.
Peakall, R. & P. E. Smouse, 2012. GenAlEx 6.5: genetic analysis in Excel Population genetic software for teaching and research-an update. Bioinformatics 28(19): 2537–2539.
Pettersen, R. A., T. A. Mo, H. Hansen & L. A. Vøllestad, 2015. Genetic population structure of Gyrodactylus thymalli (Monogenea) in a large Norwegian river system. Parasitology 142(14): 1693–1702.
Pickering, A. D. & P. Christie, 1980. Sexual differences in the incidence and severity of ectoparasitic infestation of the Brown trout, Salmo trutta L. Journal of Fish Biology 16(6): 669–683.
Pike, A. W. & J. W. Lewis, 1994. Parasit. dis. fish. Dyfed, Wales: Samara Publishing.
Plaisance, L., T. Huyse, D. T. J. Littlewood, T. A. Bakke & L. Bachmann, 2007. The complete mitochondrial DNA sequence of the monogenean Gyrodactylus thymalli (Platyhelminthes: Monogenea), a parasite of grayling (Thymallus thymallus). Molecular and Biochemical Parasitology 154(2): 190–194.
Platt, W. J. & I. M. Weis, 1985. An experimental study of competition among fungitive prairie plants. Ecology 66(3): 708–720.
Pond, S. L. K. & S. D. W. Frost, 2005. Not so different after all: a comparison of methods for detecting amino acid sites under selection. Molecular Biology and Evolution 22(5): 1208–1222.
Poulin, R., 1997. Parasite faunas of freshwater fish: the relationship between richness and the specificity of parasites. The International Journal for Parasitology 27(9): 1091–1098.
Poulin, R., 2007. Evolutionary Ecology of Parasites, 2nd ed. Princeton University Press, Princeton.
Pritchard, J. K., M. Stephens & P. Donnelly, 2000. Inference of population structure using multilocus genotype data. Genetics 155(2): 945–959.
Quinn, T. P., 2005. The behavior and ecology of Pacific salmon and trout. American Fisheries Society & University of Washington Press, Seattle.
Raeymaekers, J. A. M., T. Huyse, H. Maelfait, B. Hellemans & F. A. M. Volckaert, 2008. Community structure, population structure and topographical specialisation of Gyrodactylus (Monogenea) ectoparasites living on sympatric stickleback species. Folia Parasitologica 55(3): 187–196.
Richards, E. L., C. van Oosterhout & J. Cable, 2012. Interactions between males guppies facilitates the transmission of the monogenean ectoparasite Gyrodactylus turnbulli. Experimental Parasitology 132(4): 483–486.
Richardson, J. L., S. P. Brady, I. J. Wang & S. F. Spear, 2016. Navigating the pitfalls and promise of landscape genetics. Molecular Ecology 25(4): 849–863.
Rousset, F., 1997. Genetic differentiation and estimation of gene flow from F-statistics under isolation by distance. Genetics 145(4): 1219–1228.
Rousset, F., 2008. GENEPOP ‘ 007: a complete re-implementation of the GENEPOP software for Windows and Linux. Molecular Ecology Resources 8(1): 103–106.
Saitou, N. & M. Nei, 1987. The neighbor-joining method—a new method for reconstructing phylogenetic trees. Molecular Biology and Evolution 4(4): 406–425.
Skaala, O., 1992. Genetic population structure of Norwegian brown trout. Journal of Fish Biology 41(4): 631–646.
Slatkin, M., 1987. Gene flow and the geographic structure of natural populations. Science 236(4803): 787–792.
Soleng, A., A. B. S. Polèo, N. E. W. Alstad & T. A. Bakke, 1999. Aqueous aluminium eliminates Gyrodactylus salaris (Platyhelminthes, Monogenea) infections in Atlantic salmon. Parasitology 119: 19–25.
Song, R., W. X. Li, S. G. Wu, H. Zou & G. T. Wang, 2014. Population genetic structure of the acanthocephalan Acanthosentis cheni in anadromous, freshwater, and landlocked stocks of its fish host, Coilia nasus. Journal of Parasitology 100(2): 193–197.
Sprehn, C. G., M. J. Blum, T. P. Quinn & D. C. Heins, 2015. Landscape genetics of Schistocephalus solidus parasites in threespine stickleback (Gasterosteus aculeatus) from Alaska. PLoS ONE 10(4): 17.
Stabell, O. B., 1984. Homing and olfaction in salmonids: a critical review with special reference to the Atlantic salmon. Biol Rev Cambridge Philosophic Soc 59(3): 333–388.
Suzuki, Y. & T. Gojobori, 1999. A method for detecting positive selection at single amino acid sites. Molecular Biology and Evolution 16(10): 1315–1328.
Tajima, F., 1989. Statistical method for testing the neutral mutation hypothesis by DNA polymorphism. Genetics 123(3): 585–595.
Tamura, K., 1992. The rate and pattern of nucleotide substitution in Drosophila mitochondrial DNA. Molecular Biology and Evolution 9(5): 814–825.
Tamura, K., M. Nei & S. Kumar, 2004. Prospects for inferring very large phylogenies by using the neighbor-joining method. Proceedings of the National Academy of Sciences of the United States of America 101(30): 11030–11035.
Tamura, K., J. Dudley, M. Nei & S. Kumar, 2007. MEGA4: molecular evolutionary genetics analysis (MEGA) software version 40. Molecular Biology and Evolution 24(8): 1596–1599.
Van Leeuwen, C. H. A., K. Dalen, J. Museth, C. Junge & L. A. Vøllestad, 2018. Habitat fragmentation has interactive effects on the population genetic diversity and individual behaviour of a freshwater salmonid fish. River Research and Applications 34(1): 60–68.
Walther, B. A., P. Cotgreave, R. D. Price, R. D. Gregory & D. H. Clayton, 1995. Sampling effort and parasite species richness. Parasitol Today 11(8): 306–310.
Waples, R. S., 2004. Salmonid Insights into Effective Population Size. Oxford University Press, Oxford.
Weir, B. S. & C. C. Cockerham, 1984. Estimating F-statistics for the analysis of populaion structure. Evolution 38(6): 1358–1370.
Wright, S., 1943. Isolation by distance. Genetics 28(2): 114–138.
Acknowledgements
We thank Nanna Winger Steen and Emelita Rivera Nerli for excellent technical assistance, Haakon Hansen for assistance of the selection of genetic markers for G. thymalli, Anders Nielsen for assistance with statistics, and Finn Gregersen for assistance during the field work.
Funding
Open Access funding provided by Norwegian Institute of Bioeconomy Research. This project was mainly funded by a grant by the University of Oslo to RAP, with additional funding from the Norwegian Research Council to LAV (No. 177728).
Author information
Authors and Affiliations
Contributions
RAP, CJ, KØ, TAM and LAV designed the study. RAP collected the grayling and performed data collection. RAP and CJ have conducted the molecular analyzes and analyzed the results. RAP, CJ, KØ, TAM and LAV co-drafted the manuscript together. All authors read and approved the final version of the manuscript.
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no competing interests.
Ethical approval
The collection of grayling had permission from the authorities granted by the County Governor of Hedemark and the County Governor of Oppland. The fish was killed shortly after capture and were subjected to stress as little as possible.
Additional information
Handling editor: Christian Sturmbauer
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Pettersen, R.A., Junge, C., Østbye, K. et al. Genetic population structure of the monogenean parasite Gyrodactylus thymalli and its host European grayling (Thymallus thymallus) in a large Norwegian lake. Hydrobiologia 848, 547–561 (2021). https://doi.org/10.1007/s10750-020-04431-7
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10750-020-04431-7