BmFoxO Gene Regulation of the Cell Cycle Induced by 20-Hydroxyecdysone in BmN-SWU1 Cells
Abstract
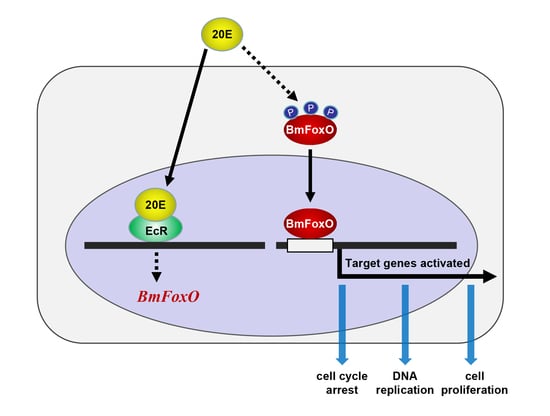
:Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Bioinformation Analysis
2.2. Cell Culture and Transient Transfections
2.3. Plasmid Construction
2.4. 20E Treatment
2.5. Quantitative Real-Time Polymerase Chain Reaction (qRT-PCR)
2.6. Flow Cytometry
2.7. BrdU Incorporation and Immunofluorescence
2.8. MTT Assay
2.9. Nuclear and Cytoplasmic Protein Extraction and Western Blot
2.10. Statistical Analysis
3. Results
3.1. Regulation of the Cell Cycle by 20E
3.2. 20E Can Regulate the Cell Cycle through BmEcR in BmN-SWU1 Cells
3.3. The BmFoxO Gene Is Necessary for Cell Cycle Regulation Induced by 20E
3.4. BmFoxO Inhibits Cell Proliferation and Causes Cell Cycle Arrest
4. Discussion
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Nijhout, H.F.; Callier, V. Developmental mechanisms of body size and wing-body scaling in insects. Annu. Rev. Entomol. 2015, 60, 141–156. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ninov, N.; Martin-Blanco, E. Changing gears in the cell cycle: Histoblasts and beyond. Fly (Austin) 2009, 3, 286–289. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Riddiford, L.M.; Hiruma, K.; Zhou, X.; Nelson, C.A. Insights into the molecular basis of the hormonal control of molting and metamorphosis from Manduca sexta and Drosophila melanogaster. Insect Biochem. Mol. Biol. 2003, 33, 1327–1338. [Google Scholar] [CrossRef] [PubMed]
- Pesch, Y.-Y.; Hesse, R.; Ali, T.; Behr, M. A cell surface protein controls endocrine ring gland morphogenesis and steroid production. Dev. Biol. 2019, 445, 16–28. [Google Scholar] [CrossRef] [PubMed]
- Ishimoto, H.; Kitamoto, T. Beyond molting—Roles of the steroid molting hormone ecdysone in regulation of memory and sleep in adult Drosophila. Fly (Austin) 2011, 5, 215–220. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, W.N.; Ma, L.; Liu, C.; Chen, L.; Xiao, H.J.; Liang, G.M. Dissecting the role of Kruppel homolog 1 in the metamorphosis and female reproduction of the cotton bollworm, Helicoverpa armigera. Insect Mol. Biol. 2018, 27, 492–504. [Google Scholar] [CrossRef] [PubMed]
- Dong, Y.; Dinan, L.; Friedrich, M. The effect of manipulating ecdysteroid signaling on embryonic eye development in the locust Schistocerca americana. Dev. Genes Evol. 2003, 213, 587–600. [Google Scholar] [CrossRef]
- Friesen, K.J.; Kaufman, W.R. Effects of 20-hydroxyecdysone and other hormones on egg development, and identification of a vitellin-binding protein in the ovary of the tick, Amblyomma hebraeum. J. Insect Physiol. 2004, 50, 519–529. [Google Scholar] [CrossRef]
- Li, Y.F.; Chen, X.Y.; Zhang, C.D.; Tang, X.F.; Wang, L.; Liu, T.H.; Pan, M.H.; Lu, C. Effects of starvation and hormones on DNA synthesis in silk gland cells of the silkworm, Bombyx mori. Insect Sci. 2016, 23, 569–578. [Google Scholar] [CrossRef]
- Rewitz, K.F.; Rybczynski, R.; Warren, J.T.; Gilbert, L.I. Developmental expression of Manduca shade, the P450 mediating the final step in molting hormone synthesis. Mol. Cell. Endocrinol. 2006, 247, 166–174. [Google Scholar] [CrossRef]
- Kovalenko, E.V.; Mazina, M.Y.; Krasnov, A.N.; Vorobyeva, N.E. The Drosophila nuclear receptors EcR and ERR jointly regulate the expression of genes involved in carbohydrate metabolism. Insect Biochem. Mol. Biol. 2019, 112, 103184. [Google Scholar] [CrossRef]
- Vafopoulou, X.; Steel, C.G. Insulin-like and testis ecdysiotropin neuropeptides are regulated by the circadian timing system in the brain during larval-adult development in the insect Rhodnius prolixus (Hemiptera). Gen. Comp. Endocrinol. 2012, 179, 277–288. [Google Scholar] [CrossRef] [PubMed]
- Fallon, A.M.; Gerenday, A. Ecdysone and the cell cycle: Investigations in a mosquito cell line. J. Insect Physiol. 2010, 56, 1396–1401. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Koyama, T.; Iwami, M.; Sakurai, S. Ecdysteroid control of cell cycle and cellular commitment in insect wing imaginal discs. Mol. Cell. Endocrinol. 2004, 213, 155–166. [Google Scholar] [CrossRef] [PubMed]
- Mitchell, N.; Cranna, N.; Richardson, H.; Quinn, L. The Ecdysone-inducible zinc-finger transcription factor Crol regulates Wg transcription and cell cycle progression in Drosophila. Development 2008, 135, 2707–2716. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mitchell, N.C.; Lin, J.I.; Zaytseva, O.; Cranna, N.; Lee, A.; Quinn, L.M. The Ecdysone receptor constrains wingless expression to pattern cell cycle across the Drosophila wing margin in a Cyclin B-dependent manner. BMC Dev. Biol. 2013, 13, 28. [Google Scholar] [CrossRef] [Green Version]
- Suren-Castillo, S.; Abrisqueta, M.; Maestro, J.L. FoxO is required for the activation of hypertrehalosemic hormone expression in cockroaches. Biochim. Biophys. Acta 2014, 1840, 86–94. [Google Scholar] [CrossRef] [Green Version]
- La, G.; Zhou, M.; Lim, J.Y.; Oh, S.; Xing, H.; Liu, N.; Yang, Y.; Liu, X.; Zhong, L. Proteomics and transcriptomics analysis reveals clues into the mechanism of the beneficial effect of electrical stimulation on rat denervated gastrocnemius muscle. Cell. Physiol. Biochem. 2019, 52, 769–786. [Google Scholar]
- Nowak, K.; Gupta, A.; Stocker, H. FoxO restricts growth and differentiation of cells with elevated TORC1 activity under nutrient restriction. PLoS Genet. 2018, 14, e1007347. [Google Scholar] [CrossRef] [Green Version]
- Ding, R.; Wang, X.; Chen, W.; Li, Z.; Wei, A.L.; Wang, Q.B.; Nie, A.H.; Wang, L.L. WX20120108, a novel IAP antagonist, induces tumor cell autophagy via activating ROS-FOXO pathway. Acta Pharmacol. Sin. 2019, 40, 1466–1479. [Google Scholar] [CrossRef]
- Cai, M.J.; Zhao, W.L.; Jing, Y.P.; Song, Q.; Zhang, X.Q.; Wang, J.X.; Zhao, X.F. 20-Hydroxyecdysone activates Forkhead box O to promote proteolysis during Helicoverpa armigera molting. Development 2016, 143, 1005–1015. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pan, M.H.; Cai, X.J.; Liu, M.; Lv, J.; Tang, H.; Tan, J.; Lu, C. Establishment and characterization of an ovarian cell line of the silkworm, Bombyx mori. Tissue Cell 2010, 42, 42–46. [Google Scholar] [CrossRef] [PubMed]
- Hossain, M.S.; Liu, Y.; Zhou, S.; Li, K.; Tian, L.; Li, S. 20-Hydroxyecdysone-induced transcriptional activity of FoxO upregulates brummer and acid lipase-1 and promotes lipolysis in Bombyx fat body. Insect Biochem. Mol. Biol. 2013, 43, 829–838. [Google Scholar] [CrossRef] [PubMed]
- Dong, Z.Q.; Chen, T.T.; Zhang, J.; Hu, N.; Cao, M.Y.; Dong, F.F.; Jiang, Y.M.; Chen, P.; Lu, C.; Pan, M.H. Establishment of a highly efficient virus-inducible CRISPR/Cas9 system in insect cells. Antivir. Res. 2016, 130, 50–57. [Google Scholar] [CrossRef] [PubMed]
- Moriyama, M.; Osanai, K.; Ohyoshi, T.; Wang, H.B.; Iwanaga, M.; Kawasaki, H. Ecdysteroid promotes cell cycle progression in the Bombyx wing disc through activation of c-Myc. Insect Biochem. Mol. Biol. 2016, 70, 1–9. [Google Scholar] [CrossRef]
- Pan, C.W.; Jin, X.; Zhao, Y.; Pan, Y.; Yang, J.; Karnes, R.J.; Zhang, J.; Wang, L.; Huang, H. AKT-phosphorylated FOXO1 suppresses ERK activation and chemoresistance by disrupting IQGAP1-MAPK interaction. EMBO J. 2017, 36, 995–1010. [Google Scholar] [CrossRef] [PubMed]
- Matsuzaki, H.; Daitoku, H.; Hatta, M.; Tanaka, K.; Fukamizu, A. Insulin-induced phosphorylation of FKHR (Foxo1) targets to proteasomal degradation. Proc. Natl. Acad. Sci. USA 2003, 100, 11285–11290. [Google Scholar] [CrossRef] [Green Version]
- Hou, L.; Cai, M.J.; Liu, W.; Song, Q.; Zhao, X.F. Small GTPase Rab4b participates in the gene transcription of 20-hydroxyecdysone and insulin pathways to regulate glycogen level and metamorphosis. Dev. Biol. 2012, 371, 13–22. [Google Scholar] [CrossRef] [Green Version]
- Saltiel, A.R.; Kahn, C.R. Insulin signalling and the regulation of glucose and lipid metabolism. Nature 2001, 414, 799–806. [Google Scholar] [CrossRef]
- Tran, H.; Brunet, A.; Grenier, J.M.; Datta, S.R.; Fornace, A.J., Jr.; Distefano, P.S.; Chiang, L.W.; Greenberg, M.E. DNA repair pathway stimulated by the forkhead transcription factor FOXO3a through the Gadd45 protein. Science 2002, 296, 530–534. [Google Scholar] [CrossRef] [Green Version]
- Huang, H.; Tindall, D.J. Dynamic FoxO transcription factors. J. Cell Sci. 2007, 120, 2479–2487. [Google Scholar] [CrossRef] [PubMed] [Green Version]




Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhang, Q.; Yang, J.; Chen, P.; Liu, T.; Xiao, Q.; Zhou, X.; Wang, L.; Long, Y.; Dong, Z.; Pan, M.; et al. BmFoxO Gene Regulation of the Cell Cycle Induced by 20-Hydroxyecdysone in BmN-SWU1 Cells. Insects 2020, 11, 700. https://doi.org/10.3390/insects11100700
Zhang Q, Yang J, Chen P, Liu T, Xiao Q, Zhou X, Wang L, Long Y, Dong Z, Pan M, et al. BmFoxO Gene Regulation of the Cell Cycle Induced by 20-Hydroxyecdysone in BmN-SWU1 Cells. Insects. 2020; 11(10):700. https://doi.org/10.3390/insects11100700
Chicago/Turabian StyleZhang, Qian, Jigui Yang, Peng Chen, Taihang Liu, Qin Xiao, Xiaolin Zhou, Ling Wang, Yanbi Long, Zhanqi Dong, Minhui Pan, and et al. 2020. "BmFoxO Gene Regulation of the Cell Cycle Induced by 20-Hydroxyecdysone in BmN-SWU1 Cells" Insects 11, no. 10: 700. https://doi.org/10.3390/insects11100700