Early Inoculation of Microbial Suspension in Suckling Piglets Affects the Transmission of Maternal Microbiota and the Associated Antibiotic Resistance Genes
Abstract
:1. Introduction
2. Materials and Methods
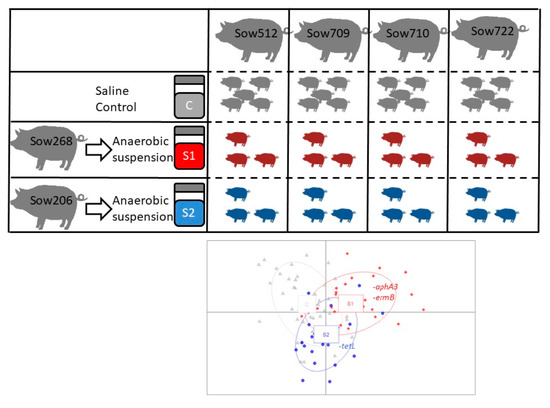
2.1. Donor Sows and Preparation of the Microbial Suspensions
2.2. Lactating Sows and Experimental Piglets
2.3. Feces Sampling and DNA Preparation
2.4. Analysis of the Antibiotic Resistance Genes via qPCR
2.5. Analysis of the Culturable Enterobacteriacae
2.6. Analysis of the Microbiota Composition Using the 16S rRNA Gene
2.7. Statistical Analysis
3. Results
3.1. The Donor and Lactating Sows Had Different Resistance Profiles
3.2. Oral Inoculation of Microbial Suspension Affected the ARGs in Piglet Feces
3.3. Oral Inoculation of Microbial Suspension Temporarily Affected the Antibiotic Resistance of Enterobacteriacae
3.4. Oral Inoculation of Microbial Suspension Affected the Vertical Transmission of Bacteria
3.5. Relationships between OTUs and ARGs
4. Discussion
4.1. Promoting Sow-to-Piglet Transmission
4.2. The Impact of Anaerobic Microbes on Antibiotic Resistance Genes
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Xiao, L.; Estelle, J.; Kiilerich, P.; Ramayo-Caldas, Y.; Xia, Z.; Feng, Q.; Liang, S.; Pedersen, A.O.; Kjeldsen, N.J.; Liu, C.; et al. A reference gene catalogue of the pig gut microbiome. Nat. Microbiol. 2016. [Google Scholar] [CrossRef] [PubMed]
- Gerzova, L.; Babak, V.; Sedlar, K.; Faldynova, M.; Videnska, P.; Cejkova, D.; Jensen, A.N.; Denis, M.; Kerouanton, A.; Ricci, A.; et al. Characterization of antibiotic resistance gene abundance and microbiota composition in feces of organic and conventional pigs from four EU countries. PLoS ONE 2015, 10, e0132892. [Google Scholar] [CrossRef] [PubMed]
- Forslund, K.; Sunagawa, S.; Kultima, J.R.; Mende, D.R.; Arumugam, M.; Typas, A.; Bork, P. Country-specific antibiotic use practices impact the human gut resistome. Genome Res. 2013, 23, 1163–1169. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chen, Y.; Hammer, E.E.; Richards, V.P. Phylogenetic signature of lateral exchange of genes for antibiotic production and resistance among bacteria highlights a pattern of global transmission of pathogens between humans and livestock. Mol. Phylogenet. Evol. 2018, 125, 255–264. [Google Scholar] [CrossRef] [PubMed]
- Aarestrup, F. Get pigs off antibiotics. Nature 2012, 486, 465–466. [Google Scholar] [CrossRef]
- Diana, A.; Manzanilla, E.G.; Calderón Díaz, J.A.; Leonard, F.C.; Boyle, L.A. Do weaner pigs need in-feed antibiotics to ensure good health and welfare? PLoS ONE 2017, 12, e0185622. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Diana, A.; Boyle, L.A.; Leonard, F.C.; Carroll, C.; Sheehan, E.; Murphy, D.; Manzanilla, E.G. Removing prophylactic antibiotics from pig feed: How does it affect their performance and health? BMC Vet. Res. 2019, 15, 67. [Google Scholar] [CrossRef] [PubMed]
- Cromwell, G.L. Why and how antibiotics are used in swine production. Anim. Biotechnol. 2002, 13, 7–27. [Google Scholar] [CrossRef]
- Abreu, R.; Rodríguez-Álvarez, C.; Lecuona, M.; Castro, B.; González, J.C.; Aguirre-Jaime, A.; Arias, Á. Increased Antimicrobial Resistance of MRSA Strains Isolated from Pigs in Spain between 2009 and 2018. Vet. Sci. 2019, 6, 38. [Google Scholar] [CrossRef] [Green Version]
- Zeitouni, S.; Kempf, I. Fitness cost of fluoroquinolone resistance in Campylobacter coli and Campylobacter jejuni. Microb. Drug Resist. 2012, 17, 171–179. [Google Scholar] [CrossRef]
- Jernberg, C.; Lofmark, S.; Edlund, C.; Jansson, J.K. Long-term impacts of antibiotic exposure on the human intestinal microbiota. Microbiology 2010, 156, 3216–3223. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jernberg, C.; Lofmark, S.; Edlund, C.; Jansson, J.K. Long-term ecological impacts of antibiotic administration on the human intestinal microbiota. ISME J. 2007, 1, 56–66. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Abecia, L.; Fondevila, M.; Balcells, J.; McEwanz, N.R. The effect of lactating rabbit does on the development of the caecal microbial community in the pups they nurture. J. Appl. Microbiol. 2007, 103, 557–564. [Google Scholar] [CrossRef] [PubMed]
- Thompson, C.L.; Wang, B.; Holmes, A.J. The immediate environment during postnatal development has long-term impact on gut community structure in pigs. ISME J. 2008, 2, 739–748. [Google Scholar] [CrossRef]
- Nurmi, E.; Rantala, M. New aspects of Salmonella infection in broiler production. Nature 1973, 241, 210–211. [Google Scholar] [CrossRef]
- Hofacre, C.L.; Johnson, A.C.; Kelly, B.J.; Froyman, R. Effect of a commercial competitive exclusion culture on reduction of colonization of an antibiotic-resistant pathogenic Escherichia coli in day-old broiler chickens. Avian Dis. 2002, 46, 198–202. [Google Scholar] [CrossRef]
- Nuotio, L.; Schneitz, C.; Nilsson, O. Effect of competitive exclusion in reducing the occurrence of Escherichia coli producing extended-spectrum beta-lactamases in the ceca of broiler chicks. Poult. Sci. 2013, 92, 250–254. [Google Scholar] [CrossRef]
- Ubeda, C.; Bucci, V.; Caballero, S.; Djukovic, A.; Toussaint, N.C.; Equinda, M.; Lipuma, L.; Ling, L.; Gobourne, A.; No, D.; et al. Intestinal microbiota containing Barnesiella species cures vancomycin-resistant Enterococcus faecium colonization. Infect. Immun. 2013, 81, 965–973. [Google Scholar] [CrossRef] [Green Version]
- Chapman, B.C.; Moore, H.B.; Overbey, D.M.; Morton, A.P.; Harnke, B.; Gerich, M.E.; Vogel, J.D. Fecal microbiota transplant in patients with Clostridium difficile infection: A systematic review. J. Trauma Acute Care Surg. 2016, 81, 756–764. [Google Scholar] [CrossRef]
- Staley, C.; Hamilton, M.J.; Vaughn, B.P.; Graiziger, C.T.; Newman, K.M.; Kabage, A.J.; Sadowsky, M.J.; Khoruts, A. Successful resolution of recurrent Clostridium difficile infection using freeze-dried, encapsulated fecal microbiota; pragmatic cohort study. Am. J. Gastroenterol. 2017. [Google Scholar] [CrossRef] [Green Version]
- Anderson, R.C.; Stanker, L.H.; Young, C.R.; Buckley, S.A.; Genovese, K.J.; Harvey, R.B.; Deloach, J.R.; Keith, N.K.; Nisbet, D.J. Effect of competitive exclusion treatment on colonization of early-weaned pigs by Salmonella serovar Choleraesuis. J. Swine Health Prod. 1999, 7, 155–160. [Google Scholar]
- Genovese, K.J.; Anderson, R.C.; Harvey, R.B.; Callaway, T.R.; Poole, T.L.; Edrington, T.S.; Fedorka-Cray, P.J.; Nisbet, D.J. Competitive exclusion of Salmonella from the gut of neonatal and weaned pigs. J. Food Prot. 2003, 66, 1353–1359. [Google Scholar] [CrossRef] [PubMed]
- Kim, L.M.; Gray, J.T.; Bailey, J.S.; Jones, R.D.; Fedorka-Cray, P.J. Effect of porcine-derived mucosal competitive exclusion culture on antimicrobial resistance in Escherichia coli from growing piglets. Foodborne Pathog. Dis. 2005, 2, 317–329. [Google Scholar] [CrossRef] [PubMed]
- Luyckx, K.; Millet, S.; Van Weyenberg, S.; Herman, L.; Heyndrickx, M.; Dewulf, J.; De Reu, K. Comparison of competitive exclusion with classical cleaning and disinfection on bacterial load in pig nursery units. BMC Vet. Res. 2016, 12. [Google Scholar] [CrossRef] [Green Version]
- Verschuren, L.M.G.; Calus, M.P.L.; Jansman, A.J.M.; Bergsma, R.; Knol, E.F.; Gilbert, H.; Zemb, O. Fecal microbial composition associated with variation in feed efficiency in pigs depends on diet and sex. J. Anim. Sci. 2018, 96, 1405–1418. [Google Scholar] [CrossRef]
- Eucast. Available online: http://www.eucast.org/ (accessed on 14 May 2015).
- Le Floc’h, N.; Knudsen, C.; Gidenne, T.; Montagne, L.; Merlot, E.; Zemb, O. Impact of feed restriction on health, digestion and faecal microbiota of growing pigs housed in good or poor hygiene conditions. Animal 2014, 8, 1632–1642. [Google Scholar] [CrossRef] [Green Version]
- Magoc, T.; Salzberg, S.L. FLASH: Fast length adjustment of short reads to improve genome assemblies. Bioinformatics 2011, 27, 2957–2963. [Google Scholar] [CrossRef]
- NCBI Sequence Read Archive. Available online: https://trace.ncbi.nlm.nih.gov/Traces/sra/sra.cgi (accessed on 7 October 2018).
- Edgar, R.C. UPARSE: Highly accurate OTU sequences from microbial amplicon reads. Nat. Methods 2013, 10, 996–998. [Google Scholar] [CrossRef]
- McMurdie, P.J.; Holmes, S. Phyloseq: An R package for reproducible interactive analysis and graphics of microbiome census data. PLoS ONE 2013, 8, e61217. [Google Scholar] [CrossRef] [Green Version]
- Jombart, T.; Devillard, S.; Balloux, F. Discriminant analysis of principal components: A new method for the analysis of genetically structured populations. BMC Genet. 2010, 11, 94. [Google Scholar] [CrossRef] [Green Version]
- Burns, A.R.; Stephens, W.Z.; Stagaman, K.; Wong, S.; Rawls, J.F.; Guillemin, K.; Bohannan, B.J. Contribution of neutral processes to the assembly of gut microbial communities in the zebrafish over host development. ISME J. 2016, 10, 655–664. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Reshef, D.N.; Reshef, Y.A.; Finucane, H.K.; Grossman, S.R.; McVean, G.; Turnbaugh, P.J.; Lander, E.S.; Mitzenmacher, M.; Sabeti, P.C. Detecting Novel Associations in Large Data Sets. Science 2011, 334, 1518–1524. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Leser, T.D.; Amenuvor, J.Z.; Jensen, T.K.; Lindecrona, R.H.; Boye, M.; Moller, K. Culture-independent analysis of gut bacteria: The pig gastrointestinal tract microbiota revisited. Appl. Environ. Microbiol. 2002, 68, 673–690. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pang, X.; Hua, X.; Yang, Q.; Ding, D.; Che, C.; Cui, L.; Jia, W.; Bucheli, P.; Zhao, L. Inter-species transplantation of gut microbiota from human to pigs. ISME J. 2007, 1, 156–162. [Google Scholar] [CrossRef]
- Jansman, A.J.M.; Zhang, J.; Koopmans, S.J.; Dekker, R.A.; Smidt, H. Effects of a simple or a complex starter microbiota on intestinal microbiota composition in caesarean derived piglets. J. Anim. Sci. 2012, 90, 433–435. [Google Scholar] [CrossRef] [Green Version]
- Gleed, P.; Sansom, B. Ingestion of iron in sows feces by piglets in farrowing crates with slotted floors. Br. J. Nutr. 1982, 47, 113–117. [Google Scholar] [CrossRef] [Green Version]
- Schmidt, B.; Mulder, I.E.; Musk, C.C.; Aminov, R.I.; Lewis, M.; Stokes, C.R.; Bailey, M.; Prosser, J.I.; Gill, B.P.; Pluske, J.R.; et al. Establishment of normal gut microbiota is compromised under excessive hygiene conditions. PLoS ONE 2011, 6, e28284. [Google Scholar] [CrossRef] [Green Version]
- Hu, J.; Ma, L.; Nie, Y.; Chen, J.; Zheng, W.; Wang, X.; Xie, C.; Zheng, Z.; Wang, Z.; Yang, T.; et al. A Microbiota-Derived Bacteriocin Targets the Host to Confer Diarrhea Resistance in Early-Weaned Piglets. Cell Host Microbe 2018, 24, 817–832. [Google Scholar] [CrossRef] [Green Version]
- Volf, J.; Polansky, O.; Varmuzova, K.; Gerzova, L.; Sekelova, Z.; Faldynova, M.; Babak, V.; Medvecky, M.; Smith, A.L.; Kaspers, B.; et al. Transient and prolonged response of chicken cecum mucosa to colonization with different gut microbiota. PLoS ONE 2016, 11, e0163932. [Google Scholar] [CrossRef]
- Achard, C.S.; Dupouy, V.; Siviglia, S.; Arpaillange, N.; Cauquil, L.; Bousquet-Mélou, A.; Zemb, O. Variability of the Ability of Complex Microbial Communities to Exclude Microbes Carrying Antibiotic Resistance Genes in Rabbits. Front. Microbiol. 2019, 10, 1503. [Google Scholar] [CrossRef]
- Kiros, T.G.; Derakhshani, H.; Pinloche, E.; D’Inca, R.; Marshall, J.; Auclair, E.; Khafipour, E.; Van Kessel, A. Effect of live yeast Saccharomyces cerevisiae (Actisaf Sc 47) supplementation on the performance and hindgut microbiota composition of weanling pigs. Sci. Rep. 2018, 8, 5315. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mu, C.; Bian, G.; Su, Y.; Zhu, W. Differential Effects of Breed and Nursing on Early-Life Colonic Microbiota and Immune Status as Revealed in a Cross-Fostering Piglet Model. Appl. Environ. Microbiol. 2019, 85. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dominguez-Bello, M.G.; De Jesus-Laboy, K.M.; Shen, N.; Cox, L.M.; Amir, A.; Gonzalez, A.; Bokulich, N.A.; Song, S.J.; Hoashi, M.; Rivera-Vinas, J.I.; et al. Partial restoration of the microbiota of cesarean-born infants via vaginal microbial transfer. Nat. Med. 2016, 22, 250–253. [Google Scholar] [CrossRef] [PubMed]
- Schokker, D.; Zhang, J.; Vastenhouw, S.A.; Heilig, H.G.H.J.; Smidt, H.; Rebel, J.M.J.; Smits, M.A. Long-Lasting Effects of Early-Life Antibiotic Treatment and Routine Animal Handling on Gut Microbiota Composition and Immune System in Pigs. PLoS ONE 2015, 10, e0116523. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Stanton, T.B.; Humphrey, S.B. Persistence of Antibiotic Resistance: Evaluation of a Probiotic Approach Using Antibiotic-Sensitive Megasphaera elsdenii Strains to Prevent Colonization of Swine by Antibiotic-Resistant Strains. Appl. Environ. Microbiol. 2011, 77, 7158–7166. [Google Scholar] [CrossRef] [Green Version]
- Holman, D.B.; Chénier, M.R. Impact of subtherapeutic administration of tylosin and chlortetracycline on antimicrobial resistance in farrow-to-finish swine. FEMS Microbiol. Ecol. 2013, 85, 1–13. [Google Scholar] [CrossRef]
- Werner, G.; Hildebrandt, B.; Witte, W. Linkage of erm(B) and aadE-sat4-aphA-3 in multiple-resistant Enterococcus faecium isolates of different ecological origins. Microb. Drug Resist. 2003, 9 (Suppl. 1), S9–S16. [Google Scholar] [CrossRef]
- Kalmokoff, M.; Waddington, L.M.; Thomas, M.; Liang, K.L.; Ma, C.; Topp, E.; Dandurand, U.D.; Letellier, A.; Matias, F.; Brooks, S.P. Continuous feeding of antimicrobial growth promoters to commercial swine during the growing/finishing phase does not modify faecal community erythromycin resistance or community structure. J. Appl. Microbiol. 2011, 110, 1414–1425. [Google Scholar] [CrossRef]
- Birkegård, A.C.; Halasa, T.; Græsbøll, K.; Clasen, J.; Folkesson, A.; Toft, N. Association between selected antimicrobial resistance genes and antimicrobial exposure in Danish pig farms. Sci. Rep. 2017, 7, 9683. [Google Scholar] [CrossRef] [Green Version]
- Looft, T.; Johnson, T.A.; Allen, H.K.; Bayles, D.O.; Alt, D.P.; Stedtfeld, R.D.; Sul, W.J.; Stedtfeld, T.M.; Chai, B.; Cole, J.R.; et al. In-feed antibiotic effects on the swine intestinal microbiome. PNAS 2012, 109, 1691–1696. [Google Scholar] [CrossRef] [Green Version]
- Swords, W.E.; Wu, C.C.; Champlin, F.R.; Buddington, R.K. Postnatal changes in selected bacterial groups of the pig colonic microflora. Biol. Neonate 1993, 63, 191–200. [Google Scholar] [CrossRef] [PubMed]
- Johnson, T.A.; Stedtfeld, R.D.; Wang, Q.; Cole, J.R.; Hashsham, S.A.; Looft, T.; Zhu, Y.G.; Tiedje, J.M. Clusters of antibiotic resistance genes enriched together stay together in swine agriculture. MBio 2016, 7, e02214–e02215. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hegstad, K.; Mikalsen, T.; Coque, T.M.; Werner, G.; Sundsfjord, A. Mobile genetic elements and their contribution to the emergence of antimicrobial resistant Enterococcus faecalis and Enterococcus faecium. Clin. Microbiol. Infect. 2010, 16, 541–554. [Google Scholar] [CrossRef] [PubMed]
- Roberts, M.C. Update on macrolide–lincosamide–streptogramin, ketolide, and oxazolidinone resistance genes. FEMS Microbiol. Lett. 2008, 282, 147–159. [Google Scholar] [CrossRef] [Green Version]
- Bayley, D.P.; Rocha, E.R.; Smith, C.J. Analysis of cepA and other Bacteroides fragilis genes reveals a unique promoter structure. FEMS Microbiol. Lett. 2000, 193, 149–154. [Google Scholar] [CrossRef] [Green Version]
- Raftis, E.J.; Forde, B.M.; Claesson, M.J.; O’Toole, P.W. Unusual genome complexity in Lactobacillus salivarius JCM1046. BMC Genom. 2014, 15, 771. [Google Scholar] [CrossRef] [Green Version]


| ARG | At 14 Days | At 4 Weeks (Weaning) | P_fried_14d | P_fried_4w | ||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Sow 512 | Sow 709 | Sow 710 | Sow 722 | Sow 512 | Sow 709 | Sow 710 | Sow 722 | |||||||||||
| S1 Piglets | Control Piglets | S1 Piglets | Control Piglets | S1 Piglets | Control Piglets | S1 Piglets | Control Piglets | S1 Piglets | Control Piglets | S1 Piglets | Control Piglets | S1 Piglets | Control Piglets | S1 Piglets | Control Piglets | |||
| aphA3 | 24 | 53 | 45 | 14 | 24 | 13 | 51 | 17 | 1.4 | 1.9 | 2.1 | 2.5 | 1.6 | 3.0 | 2.9 | 3.4 | * | |
| ermB | 0.8 | 1.9 | 0.3 | 0.3 | 0.3 | 0.4 | 1.1 | 0.7 | 1.6 | 1.8 | 3.0 | 3.4 | 2.3 | 3.1 | 2.5 | 3.5 | * | |
| aadE_2 | 4.1 | 11.9 | 4.0 | 7.6 | 1.0 | 2.4 | 6.3 | 6.9 | 2.1 | 2.2 | 2.9 | 2.9 | 2.2 | 3.0 | 1.8 | 2.7 | * | |
| cmlA1 | 439 | 19 | 62 | 0 | 801 | 501 | 0 | 0 | NA | NA | NA | NA | NA | NA | NA | NA | * | |
| dfrA16 | 0.3 | 0.2 | 0.1 | 0.0 | 0.3 | 0.1 | 0.2 | 0.0 | NA | NA | NA | NA | NA | NA | NA | NA | * | |
| ermF | 0.3 | 0.1 | 0.4 | 3.6 | 0.5 | 0.2 | 0.5 | 0.2 | 0.4 | 0.3 | 5.1 | 0.8 | 1.3 | 1.1 | 1.1 | 0.2 | * | |
| IntI1 | 61.7 | 18.2 | 9.0 | 0.1 | 111.0 | 54.4 | 23.3 | 9.0 | 4.0 | 8.9 | 63.7 | 7.0 | 3.5 | 2.8 | 28.5 | 12.5 | * | |
| is6100 a | 2 | 21 | 75 | 417 | 1 | 2 | 55 | 574 | 1.2 | 2.3 | 1.1 | 0.8 | 1.6 | 2.6 | 1.0 | 1.4 | * | |
| lnuB | 0.29 | 0.00 | 0.01 | 0.00 | 0.04 | 0.04 | 0.07 | 0.03 | 3.4 | 1.5 | 1.9 | 1.0 | 1.0 | 0.8 | 1.3 | 1.0 | * | * |
| mphB | 38.2 | 0.2 | 0.1 | 1.0 | 0.0 | 0.0 | 0.3 | 0.2 | 6.5 | 0.4 | 359.5 | 21.9 | 1.5 | 0.3 | 206.2 | 33.6 | * | |
| mphE | 4.1 | 2.5 | 11.4 | 1.4 | 0.4 | 0.3 | 2.4 | 0.6 | NA | NA | NA | NA | NA | NA | NA | NA | * | |
| npmA | 4088 | 1283 | 4819 | 6 | 492 | 15 | 4351 | 2298 | 5.7 | 6.2 | 4.4 | 4.9 | 19.3 | 9.0 | 45.3 | 57.1 | * | |
| tetQ | 0.2 | 0.1 | 0.2 | 0.7 | 0.1 | 0.1 | 0.1 | 0.0 | 0.6 | 0.5 | 1.5 | 1.0 | 1.0 | 0.9 | 0.5 | 0.3 | * | |
| tnpA b | 0.008 | 0.01 | 0.02 | 0.003 | 0.001 | 0.001 | 0.003 | 0.03 | 2.1 | 0.8 | 0.3 | 0.1 | 1.4 | 0.9 | 0.5 | 0.3 | * | |
| vanTG | 0.27 | 0.05 | 0.01 | 0.004 | 0.01 | 0.01 | 0.07 | 0.005 | 0.22 | 0.05 | 0.70 | 0.2 | 0.21 | 0.07 | 0.46 | 0.03 | * | * |
| Primer Set ID | At 14 Days | At 4 Weeks (Weaning) | P_friedman_14days | P_friedman_4weeks | ||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Sow 512 | Sow 709 | Sow 710 | Sow 722 | Sow 512 | Sow 709 | Sow 710 | Sow 722 | |||||||||||
| S2 Piglets | Control Piglets | S2 Piglets | Control Piglets | S2 Piglets | Control Piglets | S2 Piglets | Control Piglets | S2 Piglets | Control Piglets | S2 Piglets | Control Piglets | S2 Piglets | Control Piglets | S2 Piglets | Control Piglets | |||
| tetL | 0.001 | 0.011 | 0.0001 | 0.0004 | 0.003 | 0.007 | 0.003 | 0.018 | 0.05 | 0.05 | 0.02 | 0.78 | 1.90 | 1.77 | 0.01 | 0.03 | * | |
| aac6Im | 66.95 | 0.24 | 0.02 | 0.01 | 0.76 | 0.15 | 2.33 | 0.24 | NA | NA | NA | NA | NA | NA | NA | NA | * | |
| aadA | 311.85 | 69.3 | 54.75 | 51.18 | 465.4 | 83.50 | 12.08 | 4.26 | 0.28 | 0.19 | 0.15 | 0.16 | 0.40 | 0.08 | 0.62 | 0.22 | * | |
| aph2Ib | 5217.6 | 7.2 | 1.3 | 1.1 | 69.9 | 10.8 | 201.7 | 17.7 | 167 | 179 | 218 | 33.2 | 14.9 | 3.5 | 147 | 129 | * | |
| dfrA16 | 0.56 | 0.19 | 0.07 | 0.04 | 0.14 | 0.12 | 0.23 | 0.03 | NA | NA | NA | NA | NA | NA | NA | NA | * | |
| is6100 a | 65.36 | 21.3 | 1257.7 | 416.8 | 2.78 | 2.37 | 643.2 | 574.4 | 3.56 | 2.31 | 2.15 | 0.82 | 1.43 | 2.58 | 1.25 | 1.36 | * | |
| lnuC | 2.83 | 1.74 | 0.50 | 1.68 | 1.06 | 1.00 | 1.80 | 1.12 | 1.87 | 1.03 | 1.62 | 1.07 | 1.47 | 0.86 | 1.12 | 1.00 | * | |
| tetW.1 | 0.45 | 0.27 | 0.24 | 0.21 | 0.17 | 0.11 | 0.25 | 0.20 | 2.08 | 1.03 | 2.39 | 1.47 | 1.25 | 0.93 | 1.78 | 1.28 | * | * |
| Type of Relationship | ARGs or IS | Targeted Antibiotics | Linked OTU | Taxonomy of the Linked OTU |
|---|---|---|---|---|
| Linear relationship: proportionality between OTU and ARG abundances | aphA3 | Kanamycin | 131 | unclassified Clostridia |
| 2302 | unclassified Clostridiales | |||
| cepA | Cephalosporin | 6 | Bacteroides fragilis | |
| tetG | Tetracycline | 198 | unclassified Bacteroidia | |
| npmA | Aminoglycoside | 10 | unclassied Clostridiales | |
| 12,054 | unclassied Clostridia | |||
| tetM | Tetracycline | 35 | Lactobacillus | |
| lnuB | Lincosamide | 43 | Turicibacter | |
| ant6-Ib | Streptomycin | 28 | unclassified Fusobacteriaceae | |
| 120 | unclassified Clostridiales | |||
| dfrA1-sul2-intl1-aadA-aadA2-strB | Diaminopyrimidine-Sulfonamide-Aminoglycoside | |||
| blaTEM- tetR- tetA | β-lactam antibiotics-Tetracylcline | |||
| mefA-tetQ | Macrolide-Tetracycline | |||
| tetO-tet40-aadE-aphA3-tetW-ermG-ermB-lnuC | Tetracycline-Streptomycin-Kanamycin-Macrolide-Lincosamide-Streptogramin B | |||
| tet32 | tetracycline | 7 | unclassified Clostridia | |
| 4458 | unclassified Firmicutes | |||
| Intl1 | - | 51 | unclassified Clostridiales | |
| ermB | Macrolide-Lincosamide-Streptogramin B | 5 | unclassified Ruminococcaceae | |
| Co-exclusion relationship: the gene was never observed together with the OTU | ermB and ermG | Macrolide-Lincosamide-Streptogramin B | 229 | Unclassified Bacteroidales |
| 100 | Unclassified Bacteroidales | |||
| 193 | Prevotella | |||
| 868 | Clostridium_sensu_stricto | |||
| tetL | Tetracycline | 57 | unclassified Firmicutes | |
| 12 | unclassified bacterium | |||
| ermF | Macrolide-Lincosamide-Streptogramin B | 5858 | unclassified Clostridia |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Achard, C.S.; Dupouy, V.; Cauquil, L.; Arpaillange, N.; Bousquet-Melou, A.; Floc’h, N.L.; Zemb, O. Early Inoculation of Microbial Suspension in Suckling Piglets Affects the Transmission of Maternal Microbiota and the Associated Antibiotic Resistance Genes. Microorganisms 2020, 8, 1576. https://doi.org/10.3390/microorganisms8101576
Achard CS, Dupouy V, Cauquil L, Arpaillange N, Bousquet-Melou A, Floc’h NL, Zemb O. Early Inoculation of Microbial Suspension in Suckling Piglets Affects the Transmission of Maternal Microbiota and the Associated Antibiotic Resistance Genes. Microorganisms. 2020; 8(10):1576. https://doi.org/10.3390/microorganisms8101576
Chicago/Turabian StyleAchard, Caroline S., Veronique Dupouy, Laurent Cauquil, Nathalie Arpaillange, Alain Bousquet-Melou, Nathalie Le Floc’h, and Olivier Zemb. 2020. "Early Inoculation of Microbial Suspension in Suckling Piglets Affects the Transmission of Maternal Microbiota and the Associated Antibiotic Resistance Genes" Microorganisms 8, no. 10: 1576. https://doi.org/10.3390/microorganisms8101576