Abstract
Activation of cyclic GMP-AMP synthase (cGAS) through sensing cytosolic double stranded DNA (dsDNA) plays a pivotal role in innate immunity against exogenous infection as well as cellular regulation under stress. Aberrant activation of cGAS induced by self-DNA is related to autoimmune diseases. cGAS accumulates at chromosomes during mitosis or spontaneously in the nucleus. Binding of cGAS to the nucleosome competitively attenuates the dsDNA-mediated cGAS activation, but the molecular mechanism of the attenuation is still poorly understood. Here, we report two cryo-electron microscopy structures of cGAS–nucleosome complexes. The structures reveal that cGAS interacts with the nucleosome as a monomer, forming 1:1 and 2:2 complexes, respectively. cGAS contacts the nucleosomal acidic patch formed by the H2A–H2B heterodimer through the dsDNA-binding site B in both complexes, and could interact with the DNA from the other symmetrically placed nucleosome via the dsDNA-binding site C in the 2:2 complex. The bound nucleosome inhibits the activation of cGAS through blocking the interaction of cGAS with ligand dsDNA and disrupting cGAS dimerization. R236A or R255A mutation of cGAS impairs the binding between cGAS and the nucleosome, and largely relieves the nucleosome-mediated inhibition of cGAS activity. Our study provides structural insights into the inhibition of cGAS activity by the nucleosome, and advances the understanding of the mechanism by which hosts avoid the autoimmune attack caused by cGAS.
Similar content being viewed by others
Introduction
The innate immune system deploys a variety of sensors to detect signs of pathogen invasion or other cellular damage signal.1 Among them, cyclic GMP-AMP synthase (cGAS) is an important cytosolic sensor recognizing double-stranded DNA (dsDNA).2,3,4 Normally, self-DNA is strictly sequestered within the nucleus and mitochondria in eukaryotic cells. When cells are threatened by pathogen infection or cellular stress, microbial DNA or self-DNA could be present in the cytosol.5 Binding with cytosolic dsDNA activates cGAS to synthesize the secondary messenger, cyclic dinucleotide cyclic GMP-AMP (cGAMP).2,4,6,7 Recognition of cGAMP by the adaptor protein stimulator of interferon genes (STING) further recruits TANK-binding kinase (TBK1) and IκB kinase, resulting in the activation of IFN regulatory factor 3 (IRF3) and nuclear factor κB (NF-κB), which induce downstream inflammatory responses such as the production of type I interferons and other cytokines.5,6,8,9,10,11
Whereas cGAS undoubtedly plays an important role in both defense of exogenous infection and regulation of cellular environment, aberrant activation of cGAS could lead to autoimmune diseases, such as Aicardi-Goutieres Syndrome (AGS) and systemic lupus erythematosus (SLE).12,13,14,15 In particular, cGAS is activated by dsDNA with no DNA sequence specificity,7,16,17,18 therefore, cells have to develop mechanisms to tightly regulate the activity of cGAS to avoid autoimmune attack. One interesting thing is that how cGAS escapes from activation by self-DNA. Some studies suggest that cGAS is located in the cytosol during interphase, and this compartmentalization prevents its access to nuclear DNA. However, cGAS is exposed to and accumulates at chromosomes during mitosis due to the breakdown of the nuclear envelop.19,20 Meanwhile, some other studies pointed out that cGAS is located both in the nucleus and the cytosol regardless of cell cycle stages, and the nuclear cGAS is associated with chromatin.21,22,23,24 In both cases, the activity of cGAS is inhibited. Recently, a study showed that nucleosomes could competitively interact with cGAS and attenuate dsDNA-induced cGAS activation.25 This study found that during mitosis, cGAS associates with mitotic chromosomes and slowly triggers cGAS-mediated IRF3 phosphorylation, and the accumulation of the phosphorylated IRF3 in mitotic arrest leads to cell apoptosis. This intriguing finding provides an attractive and plausible resolution of the puzzle of how hosts regulate the activity of cGAS against self-DNA.
Crystal structures of the catalytic domain of cGAS alone and in complex with dsDNA have shown that the association with dsDNA makes the two lobes of cGAS closer and promotes cGAS activation.7,16,17,18,26,27,28,29 cGAS binds dsDNA in a sequence-independent manner via contacts with the sugar-phosphate backbone, and forms a minimal 2:2 complex with DNA.17,28,30 This binding mode is believed to be important for the activation of cGAS. However, the structural basis and detailed inhibitory mechanism for nucleosome-mediated attenuation of cGAS activity are still poorly understood. Here, we report two cryo-electron microscopy (cryo-EM) structures of human cGAS bound to nucleosomes, with 1:1 and 2:2 binding stoichiometry, respectively. We demonstrate that the interaction between cGAS and the nucleosome prevents cGAS from further binding to dsDNA and abolishes the dimerization of cGAS, and thus blocking the dsDNA-mediated stimulation of cGAMP synthesis. Analysis of the interfaces between cGAS and nucleosomes reveals key residues mediating the inhibition. These results reveal the structural basis for cGAS–nucleosome interaction, and should facilitate the understanding of the mechanism of avoiding autoimmune attack by cGAS.
Results
Inhibition of dsDNA-mediated cGAS activation by nucleosomes
We generated both a full-length human cGAS (cGAS-FL) and a truncated variant of human cGAS containing only the catalytic domain (cGAS-CD) (Fig. 1). The dsDNA-induced cGAS dimerization is known to be critical for cGAS activation.17 Consistent with previous reports,18 our purified cGAS-CD existed as both dimers and monomers (Supplementary information, Fig. S1a). Further, we found that the purified cGAS-FL also existed as both dimers and monomers, despite that only the cGAS-FL dimer was observed previously.18 These observations confirmed that the ligand-free cGAS proteins could assemble as dimers.18 In agreement with the previous report,18 the cGAS-FL dimer showed stronger dsDNA-induced enzyme activity than those of the cGAS-CD dimer (Supplementary information, Fig. S1b). Surprisingly, we also found that while the cGAS-CD monomer could not be activated after exposed to dsDNA, the cGAS-FL monomer still possessed the ability of cGAMP synthesis, although at a weaker level than that of the cGAS-FL dimer, but at a level comparable to that of the cGAS-CD dimer (Supplementary information, Fig. S1b). These results suggest that the N-terminal domain of cGAS (cGAS-NTD), which is lacking in cGAS-CD and has been demonstrated to bind dsDNA,31,32 helps to enhance the dsDNA-induced cGAS activation, probably through facilitate the 2:2 cGAS–dsDNA complex formation.
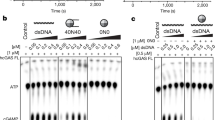
a A schematic diagram showing domains of the full-length cGAS (cGAS-FL) and the truncated cGAS (cGAS-CD) used for cryo-EM analysis. b EMSA analysis for binding of cGAS-FL and cGAS-CD in dimer or monomer states to mononucleosomes. Each lane contains 1 pmol of nucleosome reacting with 1–4 pmol of cGAS protein. The molar ratio of cGAS to the nucleosome is indicated at the top of the gel. c A bar diagram showing dose-dependent inhibition of cGAS activity by mononucleosomes. The inhibitory effects of 0–3 μM purified nucleosomes on the enzymatic activity of 5 μM dimeric cGAS against 7 μM 45 bp dsDNA were examined. Data are represented as means ± SEM (n = 3). NCP nucleosome core particle. AUC absorbance area under the curve.
Next, we prepared mononucleosomes with purified human histones and 147 bp DNA (see Method), and examined the binding affinity between cGAS and the nucleosome through electrophoretic mobility shift assay (EMSA). Both dimeric and monomeric forms of cGAS-FL and cGAS-CD shifted the nucleosomes, and the binding between cGAS-CD and the nucleosome appeared to be stronger than that between cGAS-FL and the nucleosome (Fig. 1b). These results indicate that cGAS directly interacts with the nucleosome mainly through its catalytic domain and in a manner independent of cGAS dimerization. Further, as the previous study showed that cGAS interacts with the H2A–H2B heterodimer,25 we analyzed the interaction between cGAS proteins and purified human H2A–H2B heterodimer through a pull-down assay using the poly(histidine)-tagged cGAS proteins. Both cGAS dimers and monomers bound similarly to H2A-H2B no matter whether the cGAS-NTD was present (Supplementary information, Fig. S1c), indicating a dimerization-independent interaction between cGAS-CD and the histones. To further determine the effect of cGAS activity upon binding nucleosomes, we measured the activities of cGAS dimers against dsDNA under different concentrations of nucleosomes. Consistent with previous reports,25 nucleosomes inhibited the cGAS activity in a dose-dependent manner (Fig. 1c). Moreover, we found that the activities of both the cGAS-FL and cGAS-CD dimers could be almost completely blocked by nucleosomes at a 5:3 cGAS-to-nucleosome molar ratio (Fig. 1c). The free histone H2A–H2B heterodimer could also competitively suppress the dsDNA-induced cGAS activity (Supplementary information, Fig. S1d). These observations indicate that the binding of cGAS to nucleosomes, which is mainly through cGAS-CD and in a dimerization-independent manner, accounts for the competitive inhibition of the dsDNA-induced cGAS activation by nucleosomes.
Overall structures of cGAS–nucleosome complexes
To investigate the mechanism of nucleosome-mediated inhibition of cGAS activity, we determined the cryo-EM structures of cGAS–nucleosome complexes using single particle reconstruction (Supplementary information, Fig. S2). During the cryo-EM sample preparation, we found that the complex of the cGAS-CD and nucleosomes possesses a better property for structural studies than that of the complex of cGAS-FL and nucleosomes. Moreover, since mutating the residues (KRKK, K173E/R176E/K407E/K411A) critical for dsDNA binding does not interfere with the interaction between cGAS and nucleosome as mentioned in the previous report,25 we generated the cGAS-CD with the KRKK substitution to minimize non-specific DNA binding. Interestingly, although we prepared the complexes with the cGAS-CD–KRKK dimer, our cryo-EM analysis showed that cGAS interacts with the nucleosome as a monomer (Fig. 2; Supplementary information, Fig. S2). We obtained two structures representing cGAS–nucleosome complexes at 1:1 and 2:2 molar ratios, respectively. For the structure of the 1:1 complex, which reached an overall resolution of 3.8 Å, the cGAS monomer is positioned besides the H2A–H2B patch as shown in Fig. 2a, b. A model has been built against the 1:1 complex (see Materials and Methods). Although the part of cGAS distant from the interaction region became flexible, as indicated by a relatively lower local resolution (4.7 Å) of this region (Supplementary information, Fig. S3e), the structure of cGAS around the interface between cGAS and nucleosome was well defined in the 3.8 Å map and the side chain density could be clearly observed (Supplementary information, Fig. S3a–d). For the structure of the 2:2 complex, the model of the 1:1 complex was fitted into the density map, which was determined at an overall resolution of 4.9 Å. In the 2:2 complex, one cGAS monomer binds to one nucleosome in a manner similar to that in the 1:1 complex, but the cGAS molecule is sandwiched between two nucleosomes through binding to another nucleosome via interactions with the nucleosomal DNA (Fig. 2c, d). The two nucleosomes are related by an approximately two-fold symmetry, and so are the two cGAS molecules.
a High-resolution composite cryo-EM map of the 1:1 cGAS–nucleosome complex. b Cartoon representation of the 1:1 cGAS-nucleosome complex in different views. c High-resolution composite cryo-EM map of the 2:2 cGAS–nucleosome complex. d Cartoon representation of the 2:2 cGAS–nucleosome complex in different views.
It has been demonstrated that cGAS possesses three binding sites (site A, B and C) to mediate dsDNA interaction, and all of the three sites participate in the stimulation of cGAS activity.17,18,29 As shown in the structures of cGAS–nucleosome complexes, cGAS interacts with the H2A–H2B heterodimer through the region around site B, and contacts the DNA from the adjacent nucleosome via residues within site C (Fig. 2). Thus, the dsDNA binding site B and site C are occupied by the bound nucleosomes (Fig. 2). Although site A does not directly contact the nucleosome, it is not accessible to dsDNA due to steric hindrance (Supplementary information, Fig. S4b). Thus, the absence of the interaction with dsDNA via cGAS site A would prevent the triggering of the activation loop formation as suggested by previous studies,16,28 resulting in the inhibition of the enzymatic activity of cGAS. Moreover, the cGAS dimer in both the apo state and ligand-bound forms would be captured by the bound nucleosome around the cGAS site B and disrupted into monomers (Supplementary information, Fig. S4a, b). Our data indicated that cGAS bound to nucleosome in an inactive form similar to that of the apo cGAS, with the two lobes more open compared with that of the active cGAS bound to dsDNA ligands (Supplementary information, Fig. S4c). These observations suggest that cGAS is locked to the inactive state by the binding of nucleosomes, and this binding disrupts cGAS dimerization and prevents further dsDNA binding, thus blocking dsDNA-triggered cGAS activation.
Interface between cGAS site B and the H2A–H2B heterodimer
The structures of cGAS–nucleosome complexes showed that the cGAS monomer is tightly bound to the nucleosome mainly through the acidic patch formed by the H2A–H2B heterodimer (Fig. 3a). Three loop regions protruding from the β sheet and a flanking α helix at the N-lobe of cGAS contribute to the maintenance of the complex via forming multiple electrostatic interactions and hydrogen bonds with H2A and H2B (Fig. 3b–f). The loop 1 between β2 and β3 is positioned above the nucleosomal acidic patch by the residue R236. Density of the side chain of R236, which is inserted into the acidic pocket formed by E61 and E64 from H2A, was clearly observed in the map (Fig. 3c). The loop 2 between α3 and β4 is also well resolved in our structures (Fig. 3d), whereas this loop is disordered18,26,29,33 or shifted away17,18,30 in several reported cGAS structures in ligand-free or dsDNA-bound states (Supplementary information, Fig. S5a), suggesting that the loop 2 is stabilized by the interaction between cGAS and the nucleosome. R255 within the loop 2 binds in the acidic cavity formed by E61, D90 and E92 from H2A, resembling the common binding mode of arginine residues to the acidic patch in other nucleosome binding proteins (Fig. 3d; Supplementary information, Fig. S5c–j).34,35,36,37,38,39 Furthermore, acidic patch binding by residues K254 and K258 may be responsible for stabilizing the loop 2 in the complex with the nucleosome (Fig. 3d). Interestingly, our density map shows that the side chain of H2A R71 projects toward the residues S328 and S329 within the loop protruding from β7 (loop 3) (Fig. 3e). The flanking helix α5 is also positioned adjacent to the H2A–H2B heterodimer. Although the density of this helix is relatively disordered, it is in a position to form potential contacts with H2B D51 through the side chains of R349 and R353, and interacts with H2A T76 via the side chains of K350 (Fig. 3f). Besides the above described interactions between cGAS site B and the H2A–H2B heterodimer, potential contacts between several positive charged residues from the flanking helices and nucleosomal DNA may also help to stabilize the cGAS–nucleosome complex (Supplementary information, Fig. S5b).
a Location of the cGAS binding site on the acidic nucleosomal surface. The histone octamer is shown as electrostatic surface representation. Red represents negative charge, and blue represents positive charge. b Overview of the cGAS site B bound to the H2A–H2B heterodimer. c–f Detailed view of the interaction between cGAS and H2A–H2B heterodimer formed by Loop 1 (c), Loop 2 (d), Loop 3 (e) and α5 (f) from cGAS site B. Residues at the interface are shown as sticks. Densities of the side chains of the residues from the interaction regions are shown as blue meshes based on our 3.8 Å map. The side chains of the residues from the helix α5 of cGAS that may form potential interactions are also indicated (f), although their densities could not be well resolved.
Binding sites within cGAS site B critical for nucleosome-mediated cGAS inhibition
To determine the binding specificity of cGAS to nucleosomes, we separately mutated all of the cGAS residues within site B that may contact with the HA2–H2B heterodimer, based on analysis of our structures (Supplementary information, Table S1), and examined their binding ability to both the H2A–H2B and the nucleosome through pull-down assays or EMSA. All the mutations were generated within cGAS-CD. From the pull-down results (Fig. 4a), we found that substitutions of either R236A in loop 1 or R255A in loop 2 almost totally blocked the interaction between cGAS and H2A-H2B. Both mutations of R353A from α5 and K254A from loop 2 largely impaired the binding ability of cGAS with H2A-H2B. The S329A mutant in loop 3 and the R349A mutant in α5 partially weakened the interaction. Other mutants K258A, S328A, K327A only slightly diminished the binding between cGAS and H2A–H2B, while the K350A mutant showed no effect on the interaction. Consistently, results from EMSA showed that R236A and R255A mutants largely weakened the mobility shift compared to that of wild type (WT) cGAS, and K254A and K258A also partially impaired the binding between cGAS and the nucleosome (Fig. 4b). Other mutants shifted the nucleosomes similarly to WT cGAS (Supplementary information, Fig. S6a–c). Because the presence of trace amount of free dsDNA in the purified nucleosome sample, we compared the binding abilities of cGAS-CD toward the nucleosome and free dsDNA. In agreement with the results from a previous study,25 the nucleosome shifted with cGAS much more efficiently than with free dsDNA of different lengths (Supplementary information, Fig. S6e). Thus the results from the EMSA data reflect the different binding abilities of WT cGAS-CD or cGAS-CD mutants toward the nucleosome. Moreover, we generated a K394A mutation, which is located at the cGAS dimer interface and it blocked cGAS dimerization as previously reported.17 The results further demonstrated that dimerization of cGAS is not required for cGAS binding with H2A-H2B (Fig. 4a), as well as interaction with the nucleosome (Supplementary information, Fig. S6d).
a Pull-down assays showing the effects of cGAS-CD mutants on H2A-H2B binding. MBP-His-tagged WT cGAS-CD and cGAS-CD mutants in monomer state were incubated with no-tagged H2A-H2B and NI-beads for 2 h. The bound proteins were analyzed by coomassie blue staining. b EMSA data for binding of cGAS-CD mutants to mononucleosomes. The monomeric cGAS-CD proteins were used in the EMSA experiments. Each lane contains 1 pmol of nucleosome reacting with 1–4 pmol of cGAS protein. The molar ratio of cGAS to the nucleosome is indicated at the top of the gel. c The cGAMP production level of 5 μM WT cGAS-CD or cGAS-CD mutants in dimer state against 7 μM dsDNA with or without 3 μM mononucleosomes. Data are represented as means ± SEM (n = 3). NCP nucleosome core particle. AUC absorbance area under the curve.
Then we analyzed the effect of these mutations on inhibition of cGAS activity by nucleosomes (Fig. 4c). Since the mutations are located around the DNA binding site B, most of the mutants showed relatively weaker dsDNA-mediated cGAS activation compared to that of WT cGAS. Consistent with the results from EMSA and binding assays, both mutations of R236A and R255A rescued the dsDNA-induced cGAS activation from the nucleosome-mediated inhibition, and the production level reached 80%–90% of that in the control without nucleosome treatment in both samples. Binding with nucleosomes still blocked the cGAS activation for the mutations K254A, K258A, K327A, S328A and S329A. The effects of R349A, K350A and R353A mutations could not be reliably judged because of their extremely low enzyme activities. These observations demonstrated that residues R236 from loop 1 and R255 from loop 2 are the key determinants contributing to the nucleosome-mediated prevention of cGAS activity through binding with the acidic patch formed by H2A–H2B heterodimer.
Interface between cGAS site C and the nucleosomal DNA
The structure of the 2:2 cGAS–nucleosome complex showed that each cGAS monomer interacts with the DNA from the opposite nucleosome through the DNA binding site C (Fig. 5a), which is recently reported to contact a third dsDNA and thus plays an important role in stimulating cGAS activity.18 The α-region, the KRKR-loop and the KKH-loop from site C that mediates interaction between cGAS and dsDNA are also employed in binding nucleosomal DNA through positive-charged residues within the three regions (Fig. 5b, c). Although the densities of side chains within site C could not be resolved because of the relatively lower resolution in the region, we mutated the cGAS residues in these regions that have the potential to interact with nucleosomal DNA (Supplementary information, Table S1), and examined the nucleosome-binding abilities of these mutants through EMSA. Mutations in all three regions significantly affected the mobility shift (Fig. 5d). Compared to the WT cGAS, the R300A/K301A/R302A mutation from site C completely blocked the formation of the larger complex, but shifted the nucleosomes to two lower bands, which may represent the cGAS-nucleosome complex in a 1:1 (with only one cGAS bound to one H2A–H2B heterodimer from the nucleosome) and 2:1 manner (with two cGAS bound to the H2A–H2B heterodimers at both sides of the nucleosome), respectively. The other two mutations (K282A/K285A and K427A/K428A) also greatly inhibited the formation of the larger complexes. These results suggest that the specific interactions via cGAS site C mediate the binding of cGAS to the symmetry-related nucleosome, which contributes to the formation of the 2:2 cGAS–nucleosome complex.
a Overview of the cGAS site C bound to the DNA from the adjacent nucleosome at the opposite side. b Detailed view of the interaction between cGAS site C and the nucleosomal DNA. c The electrostatic surface representation of the cGAS site C interface. Red represents negative charge, and blue represents positive charge. d EMSA analysis for binding of the cGAS-CD mutants at site C to mononucleosomes. The monomeric cGAS-CD proteins were used in the EMSA experiments. Each lane contains 1 pmol of nucleosome reacting with 1–4 pmol of cGAS protein. The molar ratio of cGAS to the nucleosome is indicated at the top of the gel. NCP, nucleosome core particle.
We further determined the effect of the mutations from site C on the nucleosome-mediated inhibition of cGAS activity and found that nucleosomes could still block the dsDNA-induced enzyme activation by all of the mutations under a molar ratio of 5:3 (Supplementary information, Fig. S7). Note that although mutations from site C prevented the formation of larger complexes, these mutants could also shift nucleosomes to lower bands efficiently (Fig. 5d). Thus, cGAS mutants at site C and nucleosomes could still interact with each other through other regions, probably the cGAS site B and the nucleosomal H2A–H2B heterodimer, which plays a critical role in the inhibition of cGAS activity by the bound nucleosomes.
Higher-order cGAS–nucleosome complex
During the cryo-EM sample screening, we observed a small amount of ladder-shaped particles indicating the cGAS–nucleosome complex may form a higher-order assembly (Supplementary information, Fig. S2f). After multiple rounds of classifications, we successfully obtained a density map with a resolution of 20 Å that representing cGAS bound to nucleosomes forming a 4:3 complex (Fig. 6a). The structures of the 1:1 and 2:2 cGAS–nucleosome complexes could be well fitted into the map, resembling three twisted hamburgers connected by additional sandwich layers (Fig. 6b–d). Two molecules of the cGAS monomer is bound symmetrically to the H2A–H2B heterodimers at both sides of the middle nucleosome, and the cGAS site C interacting with the adjacent nucleosomal DNA contributes to further binding of the other two cGAS–nucleosome complexes (Fig. 6b–d). These observations suggest that the cGAS proteins could form higher-order complexes with the nucleosome, and the higher-order complexes may further regulate the inhibition of cGAS activity.
a–d cryo-EM map and structure of higher-order cGAS-nucleosome complex in a 4:3 manner. The models of cGAS and nucleosome are fitted into the map using UCSF chimera. The model of the 1:1 cGAS–nucleosome complex on the top is colored blue, the model of the 2:1 cGAS–nucleosome complex in the middle is colored orange and the model of the 1:1 cGAS–nucleosome complex at the bottom is colored pink.
Discussion
The activation of cGAS is induced by the interaction with dsDNA ligands and dimerization of cGAS to form a minimal 2:2 complex. Since cGAS binds to dsDNA with no sequence specificity, aberrant activation of cGAS by endogenous DNA has been related to autoimmune diseases and tumors. Strict regulation of the catalytic activity is needed for the cytosolic cGAS in mitosis as well as the nuclear cGAS, which have many opportunities to contact the host DNA from chromatin. In this study, we used structural data to demonstrate that nucleosomes could not stimulate cGAS activity, but block cGAS activation through interacting with cGAS monomer and thus interfering dsDNA binding as well as cGAS dimerization. Therefore, whether in the case that cGAS accumulated to the chromosomes during mitosis, or that cGAS may be located in nucleus, the nucleosomes competitively capture cGAS to inhibit the dsDNA-induced activation and thus prevent autoimmune responses. Moreover, nucleosomes may also protect self-DNA from recognition by cGAS through tight association of the nucleosomal DNA with the histone octamers. However, we suggest that if cGAS proteins are abundant under special circumstances, they may be still able to be activated by the linker DNA within the nucleosome-free regions or other free DNA appeared in the cytosol. Thus, as previously reported, the dsDNA-induced cGAS activation was largely inhibited by nucleosomes in normal mitosis, but during mitosis arrest the mitotic chromosomes could stimulate cGAMP synthesis slowly, which led to the accumulation of phosphorylated IRF3 and further promoted mitotic apoptosis.25 Other cases were also reported to cause chromatin-related cGAS activation. For instance, cGAS is localized to the micronuclei generated by genome instability, and activated by the chromatin DNA from the micronuclei triggering downstream innate immune responses.40
According to the structures of cGAS–nucleosome complexes, we demonstrated that both the residues R236 and R255 are critical for binding of cGAS to the nucleosomal acidic patch formed by H2A and H2B, and play an important role in the inhibitory effect of nucleosomes on cGAS activity. A recent study also found that the residues R236 and R255 are pivotal for cGAS nuclear tethering and inhibition of self-DNA triggered activation by analyzing the nuclear tethering and cGAMP production in cells.22 Note that both R236 and R255 are completely conserved, indicating a common inhibitory mechanism of nucleosomes to block cGAS activation against endogenous DNA among different species. While this work was prepared for the revised manuscript, several structural studies of the cGAS–nucleosome complex were published online,41,42,43,44,45 which reached conclusions consistent with ours. Interestingly, human cGAS could bound to nucleosome forming 2:2 or higher-order complex as seen in our and the other two studies,41,44 but mouse cGAS only interacts with nucleosome in a 1:1 manner.42,43,45 The basic residues from the site C which contact the nucleosomal DNA from the symmetrically related nucleosome help to maintain the oligomerization of the human cGAS–nucleosome complex. Lack of protein sequence conservation within the site C region of human and mouse cGAS may account for this difference. Several basic residues in the KRKR-loop and KKH-loop of human cGAS that were demonstrated critical for the formation of higher-order complex in our study were substituted by acidic residues in mouse cGAS. It is interesting to investigate the biological functions of the higher-order complex in further studies, which seems unique to human cells.
Besides the C-terminal catalytic domain, cGAS contains an N-terminal domain that contributes to ligand DNA interaction, DNA-induced condensation and cGAS localization.21,31,32 Here we found that the NTD helps to improve the dsDNA-mediated stimulation of cGAS activity, indicating the importance of the NTD in regulation of cGAS activation. Moreover, other studies also reported that NTD regulates the activity of cGAS.18,31,46,47 For instance, GTPase-activating protein SH3 domain-binding protein 1 (G3BP1) promotes efficient DNA-mediated cGAS activation via binding to the cGAS-NTD.46 It is interesting to find out the position and function of NTD while cGAS-FL bound to the nucleosome, and whether the NTD contributes to the nucleosome-mediated inhibition of cGAS activity or helps further compaction of the nucleosomes through binding with the nucleosomal DNA. A recent study found that the non-enzymatic cGAS-NTD is responsible for the enrichment of cGAS on centromeres and LINE DNA repeats in the nucleus, as well as the limited activation of the nuclear cGAS.21 These chromosomal loci are known to be enriched with nucleosomes and they can form higher order chromatin structure. It is possible that the high nucleosome density and/or their specific spatial arrangement may facilitate the enrichment of cGAS, especially in view of our observation that cGAS can form higher oligomers with the nucleosome. Interestingly, they also found that endogenous cGAS did not activate the innate immune response, but overexpressed cGAS resulted in limited activation which is at least 200-fold less active compared to the induction by exogenous cytosolic DNA.21 Furthermore, nuclear tethering of cGAS should be released to sense exogenous DNA when it is localized in the cytosol during interphase. Since cGAS-CD interacts with nucleosomes with strong binding affinity,22,25 the mechanism by which cGAS is separated from nucleosomes is still unknown. It is possible that other cellular factors may help this process, for example, through contacting the regulatory cGAS-NTD. Further studies are needed to answer these questions.
In summary, our structures of cGAS–nucleosome complexes provide the structural basis and inhibitory mechanism of nucleosomes to inhibit the dsDNA-induced cGAS activation. The structures obtained here should help understanding the strategy by which hosts regulate cGAS activation to avoid autoimmune diseases.
Materials and methods
Protein expression and purification
Sequence encoding the human cGAS (UniProtKB accession Q8N884) was optimized for synthesis. The sequences corresponding to full-length (residues 1–522) and truncated (residues 157–522) cGAS were inserted into a modified pET22b vector (Novagen), in which cGAS contained an N-terminal MBP-His tag followed by a prescission protease cleavage site. Mutant proteins were constructed by site-directed mutagenesis based on the truncated cGAS-CD (157–522), respectively. All proteins were expressed in E.coli BL21 (DE3) strain. The cells were grown at 37 °C until the OD600 reached 0.6–0.8. The cells were then induced by addition of 0.3 mM isopropyl β-D-1-thiogalactopyranoside (IPTG) at 16 °C for 18 h.
Proteins were purified by Ni-NTA affinity column. The MBP-His tag was removed by prescission protease cleavage in a buffer containing 20 mM Hepes, pH 7.5, 400 mM NaCl, 10% glycerol, 1 mM DTT at 4 °C overnight. For the WT protein samples, the digested samples were further purified by a Heparin column (GE Healthcare) and then eluted with a linear gradient of 0.4–1.0 M NaCl in 20 mM Hepes buffer (pH 7.5), 10% glycerol and 1 mM DTT. For the mutant protein samples, the digested samples were further purified by a Heparin column and then eluted with linear gradient of 0.3–1.0 M NaCl in the same buffer. The two peaks from the Heparin chromatography were collected separately and further purified by size exclusion chromatography using a Superdex 200 increase 10/300 column (GE Healthcare) with a buffer containing 20 mM Hepes, pH7.5, 400 mM NaCl and 1 mM DTT, respectively. The results of gel-filtration chromatography and SDS-PAGE analysis indicated that the first peak eluted from the Heparin column is the cGAS protein in monomer state and the second peak is the cGAS protein in dimer state. For the cGAS proteins containing the MBP-His tag in the pull-down assays, the protein samples were not treated with protease digestion and purified the same as mentioned above.
Preparation of cGAS–nucleosome complex
Recombinant human histones H2A, H2B, H3, and H4 were expressed from pET3a plasmids in E.coli BL21 (DE3) as inclusion bodies and purified and reconstituted into histone octamers as described previously.48 Excessive histone H2A/H2B dimer was separated from octamers by gel filtration and pooled for pulldown experiments. Plasmid containing 12 tandem repeats of 147 bp “601” DNA49 was digested with EcoRV. The completely digested DNA fragments were separated from vector fragments using PEG precipitation following a final ethonal precipitation step. For cryo-EM sample preparation, a 40 bp linker was attached on the 3′ of “601” sequence corresponding to 601-GCATGTATTGAACAGCGACCTTGCCGGTGCCAGTCGGGATATC (EcoRV site underlined). For nucleosome reconstitution, histone octamer and DNA were mixed with a molar ratio of DNA: octamer = 1:1.1, and dialyzed against a gradient of decreasing salt concentration from 2 M KCl, 20 mM Hepes, pH 7.5, 1 mM DTT to 200 mM KCl, 20 mM Hepes, pH 7.5, 1 mM DTT, as described previously.48 Reconstituted nucleosomes were purified by DEAE-5PW exchange chromatography (Tosoh Corporation) to remove free DNA. The nucleosomes eluted were pooled and dialyzed to 20 mM Hepes, pH 7.5 and adjusted to corresponding salt concentration for future use.
Human cGAS catalytic domain with KRKK mutation was mixed with purified nucleosomes with molar ratio of cGAS:NCP = 2:1, and dialyzed to 200 mM KCl, 20 mM Hepes, pH 7.5, 1 mM DTT before loading onto a superdex 200 10/300 column (GE Healthcare) previously equilibrated with dialysis buffer. Unbound cGAS was separated from cGAS–NCP complex in different peaks.
Cryo-EM sample preparation and data collection
The complex samples with a nucleic acid concentration of 200 ng/μL were incubated with Glutaraldehyde solution (Sigma) at a final concentration of 0.25% for 30 min on ice for cross-linking reaction terminated by adding a final concentration of 90 mM Tris (pH 7.5). Holey carbon grids (GIG, 300 mesh, R2/1) were glow-discharged for 1 min before use. 3 μL of sample was placed onto the grid, incubated for 15 s and blotted for 4 s at 10 °C in 75% humidity, and then plunge-frozen in liquid ethane using an automatic plunge freezer (Leica EM GP2).
Cryo-EM single-particle data collection was carried out through the beam-image shift data collection method recently developed by our research group50 using a 200 kV FEI Tecnai Arctica microscope equipped with a K2 camera (Gatan). Images were recorded at a defocus range of −1.5 μm to −2.0 μm with a pixel size of 1.0 Å. The exposure time was 5.12 s, with a total exposure dose of ~50 electrons per Å2 over 32 frames.
Data processing
All movie stacks were corrected for beam-induced motion using MotionCor2.51 The parameter of the contrast transfer function on each micrograph was determined by the program CTFFIND4.52 Then the data were further processed in the software RELION 2.1.53 A subset of protein particles was manually picked and processed for reference-free 2D classification. Six class-averaged images were selected as references for particle auto-picking. A total of 1,955,418 particles were auto-picked and extracted from 7270 micrographs and processed for several rounds of reference-free 2D classifications. 1,060,637 particles were selected for subsequent 3D classification, which were classified into ten classes using the density map generated by the structure of the single nucleosome (PDB: 5AV5)54 as the initial model. Class 7 from the 3D classification containing 191,718 particles, represents the cGAS-nucleosome monomer with cGAS bound to nucleosome in a 1:1 manner, while class 9 from the 3D classification containing 107,374 particles, is the cGAS–nucleosome dimer with cGAS bound to nucleosome in a 2:2 manner. For the cGAS–nucleosome monomer, the particles from 3D auto-refinement were subtracted to remove most parts of the nucleosome, and the subtracted particles containing the cGAS and bound H2A–H2B were further processed for no-alignment 3D classification with a soft edged mask. 133,590 particles from the good class were selected for focused refinement and generated a map of cGAS bound with H2A-H2B at a resolution of 4.7 Å (monomer-subtracted). The particles were further re-extracted from the micrographs and used for 3D auto-refinement, generating a map of the cGAS-nucleosome monomer at 3.8 Å resolution (monomer-overall). For the cGAS–nucleosome dimer, the particles were subjected to 3D auto-refinement with C2 symmetry, yielding a 4.9 Å density map (dimer-overall). To further improve the density map, the C2 symmetry was expanded and a no-alignment classification was applied on the cGAS–nucleosome monomer with particle subtraction of the signal of the other cGAS–nucleosome monomer. The good class was selected and subjected for further focused refinement, and the resolution of the cGAS–nucleosome monomer from the 2:2 complex (dimer-subtracted) was improved to 4.3 Å using 82,617 particles. All the resolutions were estimated according to the gold-standard Fourier shell correlation with the 0.143 criterion, and the local resolution of the maps were calculated using ResMap.55
Model building and refinement
The models derived from the crystal structures of apo human cGAS (PDB: 4LEW)17 and human mononucleosome (PDB: 5AV5)54 were fitted into the cryo-EM density maps using UCSF chimera56 and manually adjusted in Coot.57 Then the models were further refined using real-space refinement in Phenix,58 and manually adjusted in Coot iteratively until no further improvement could be achieved. For the model building of cGAS protein in the 1:1 cGAS–nucleosome complex, the initial model of human cGAS was fitted, manually adjusted and refined based on the 4.7 Å cryo-EM map (monomer-subtracted), and the side chains of the amino acids from the interacting regions were further manually adjusted and refined according to the 3.8 Å cryo-EM map of the overall monomer. For the model building of the 2:2 cGAS-nucleosome complex, the structure of cGAS–nucleosome monomer was used as initial model and further fitted into the 4.3 Å cryo-EM map obtained from the subtracted dimer particles. Then the model was fitted into the 4.9 Å cryo-EM map of cGAS-nucleosome dimer. The refinement statistics of the models are summarized in Table 1. Analysis of the structures and generation of the figures representing the structural features were made using UCSF chimera56 and PyMOL (http://pymol.org).
Pull-down assays
MBP-His-tagged cGAS proteins (1 μM) or free MBP-His (1 μM, control) were mixed with no-tagged H2A–H2B heterodimers (2 μM), and incubated with Ni-beads at 4 °C for 2 h in the binding buffer (20 mM Tris-HCl pH 7.5, 500 mM NaCl, 20 mM imidazole, 0.3% (v/v) Tween-20, 5% (v/v) glycerol) with a final volume of 400 μl. The beads were washed five times with the washing buffer containing 30 mM imidazole to remove the unbound proteins. The bound proteins were then eluted using the eluting buffer containing 500 mM imidazole and analyzed by SDS-PAGE. The washing buffer and eluting buffer are the same as the binding buffer, except for the different concentrations of imidazole.
EMSA
Recombinant human mononucleosomes were incubated with full length or catalytic domain of human cGAS in EMSA buffer (50 mM Hepes, pH 7.5, 150 mM NaCl, 1 mM DTT) for 30 min at 4 °C, and further analyzed by 6% polyacrylamide, 0.25× TBE PAGE. Each lane contained 1 pmol of nucleosome reacting with indicated pmol of cGAS protein. Gels were stained by SYBR-gold to visualize DNA and loss of free NCP bands were used to indicate protein binding. The inputs of the EMSA experiments were examined by SDS-PAGE and coomassie blue staining.
cGAMP production assays
The production level of the cGAMP synthesized in the cGAS activity assay was measured using anion-exchange chromatography as previously described.17,30 5 μM purified cGAS proteins were incubated with 7 μM 45 bp dsDNA in a total of 100 μL reaction buffer (20 mM Tris, pH 7.5, 150 mM NaCl, 10 mM MgCl2, 2 mM GTP, 2 mM ATP, 1 mM DTT, 10% glycerol) at 37 °C for different time points and stopped by addition of EDTA with a final concentration of 5 μM. The reaction mixtures were centrifuged and the samples in the supernatant were diluted by 15-fold using the low salt buffer (50 mM Tris pH 8.5, 0 M NaCl) and loaded into a QHP exchange column (GE healthcare, 1 ml). The cGAMP product, the remaining GTP and ATP were eluted separately from the anion-exchange chromatography with a gradient of 0–0.5 M NaCl (50 mM Tris, pH 8.5). The absorbance area under the curve (AUC) of cGAMP at 280 nm calculated by peak integration was kept representing the production level of cGAMP. For the assays containing nucleosomes, different concentrations of nucleosomes reconstituted from 147 bp DNA were incubated with 5 μM cGAS proteins at 4 °C for 1 h in the buffer same as mentioned above, and then 7 μM 45 bp dsDNA were added into the mixtures, respectively. The samples were incubated at 37 °C for 2.5 h and analyzed by the QHP chromatography. The 45 bp dsDNA was designed as previous reports:17,18 forward sequence, 5′-TACAGATCTACTAGTGATCTATGACTGATCTGTACATGATCTACA-3′; reverse sequence, 5′-TGTAGATCATGTACAGATCAGTCATAGATCACTAGTAGATCTGTA-3′.
Data availability
The cryo-EM density maps have been deposited in the Electron Microscopy Data Bank (EMDB) under accession numbers: EMDB-30339 (monomer-overall and monomer-subtract), EMDB-30340 (dimer-overall and dimer-subtract). The atomic coordinates have been deposited in the Protein Data Bank (PDB) under accession codes: 7CCQ (monomer) and 7CCR (dimer).
References
Wu, J. & Chen, Z. J. Innate immune sensing and signaling of cytosolic nucleic acids. Annu. Rev. Immunol. 32, 461–488 (2014).
Sun, L., Wu, J., Du, F., Chen, X. & Chen, Z. J. Cyclic GMP-AMP synthase is a cytosolic DNA sensor that activates the type I interferon pathway. Science 339, 786–791 (2013).
Li, X. D. et al. Pivotal roles of cGAS-cGAMP signaling in antiviral defense and immune adjuvant effects. Science 341, 1390–1394 (2013).
Wu, J. et al. Cyclic GMP-AMP is an endogenous second messenger in innate immune signaling by cytosolic DNA. Science 339, 826–830 (2013).
Cai, X., Chiu, Y. H. & Chen, Z. J. The cGAS-cGAMP-STING pathway of cytosolic DNA sensing and signaling. Mol. Cell 54, 289–296 (2014).
Ablasser, A. et al. cGAS produces a 2’-5’-linked cyclic dinucleotide second messenger that activates STING. Nature 498, 380–384 (2013).
Gao, P. et al. Cyclic [G(2’,5’)pA(3’,5’)p] is the metazoan second messenger produced by DNA-activated cyclic GMP-AMP synthase. Cell 153, 1094–1107 (2013).
Ishikawa, H. & Barber, G. N. STING is an endoplasmic reticulum adaptor that facilitates innate immune signalling. Nature 455, 674–678 (2008).
Gao, P. et al. Structure-function analysis of STING activation by c[G(2’,5’)pA(3’,5’)p] and targeting by antiviral DMXAA. Cell 154, 748–762 (2013).
Tanaka, Y. & Chen, Z. J. STING specifies IRF3 phosphorylation by TBK1 in the cytosolic DNA signaling pathway. Sci. Signal. 5, ra20 (2012).
Fitzgerald, K. A. et al. IKKepsilon and TBK1 are essential components of the IRF3 signaling pathway. Nat. Immunol. 4, 491–496 (2003).
Crow, Y. J. & Manel, N. Aicardi-Goutières syndrome and the type I interferonopathies. Nat. Rev. Immunol. 15, 429–440 (2015).
Gao, D. et al. Activation of cyclic GMP-AMP synthase by self-DNA causes autoimmune diseases. Proc. Natl. Acad. Sci. USA 112, E5699–E5705 (2015).
Gray, E. E., Treuting, P. M., Woodward, J. J. & Stetson, D. B. Cutting edge: cGAS is required for lethal autoimmune disease in the Trex1-deficient mouse model of Aicardi-Goutières syndrome. J. Immunol. 195, 1939–1943 (2015).
Xiao, N. et al. cGAS activation causes lupus-like autoimmune disorders in a TREX1 mutant mouse model. J. Autoimmun. 100, 84–94 (2019).
Civril, F. et al. Structural mechanism of cytosolic DNA sensing by cGAS. Nature 498, 332–337 (2013).
Li, X. et al. Cyclic GMP-AMP synthase is activated by double-stranded DNA-induced oligomerization. Immunity 39, 1019–1031 (2013).
Xie, W. et al. Human cGAS catalytic domain has an additional DNA-binding interface that enhances enzymatic activity and liquid-phase condensation. Proc. Natl. Acad. Sci. USA 116, 11946–11955 (2019).
Yang, H., Wang, H., Ren, J., Chen, Q. & Chen, Z. J. cGAS is essential for cellular senescence. Proc. Natl. Acad. Sci. USA 114, E4612–E4620 (2017).
Harding, S. M. et al. Mitotic progression following DNA damage enables pattern recognition within micronuclei. Nature 548, 466–470 (2017).
Gentili, M. et al. The N-terminal domain of cGAS determines preferential association with centromeric DNA and innate immune activation in the nucleus. Cell Rep. 26, 2377–2393.e13 (2019).
Volkman, H. E., Cambier, S., Gray, E. E. & Stetson, D. B. Tight nuclear tethering of cGAS is essential for preventing autoreactivity. Elife 8, e47491 (2019).
Jiang, H., Xue, X. & Panda, S. Chromatin-bound cGAS is an inhibitor of DNA repair and hence accelerates genome destabilization and cell death. EMBO J. 38, e102718 (2019).
Liu, H. et al. Nuclear cGAS suppresses DNA repair and promotes tumorigenesis. Nature 563, 131–136 (2018).
Zierhut, C. et al. The cytoplasmic DNA sensor cGAS promotes mitotic cell death. Cell 178, 302–315.e23 (2019).
Kato, K. et al. Structural and functional analyses of DNA-sensing and immune activation by human cGAS. PLoS One 8, e76983 (2013).
Kranzusch, P. J., Lee, A. S., Berger, J. M. & Doudna, J. A. Structure of human cGAS reveals a conserved family of second-messenger enzymes in innate immunity. Cell Rep. 3, 1362–1368 (2013).
Zhang, X. et al. The cytosolic DNA sensor cGAS forms an oligomeric complex with DNA and undergoes switch-like conformational changes in the activation loop. Cell Rep. 6, 421–430 (2014).
Zhou, W. et al. Structure of the human cGAS-DNA complex reveals enhanced control of immune surveillance. Cell 174, 300–311.e11 (2018).
Andreeva, L. et al. cGAS senses long and HMGB/TFAM-bound U-turn DNA by forming protein-DNA ladders. Nature 549, 394–398 (2017).
Du, M. & Chen, Z. J. DNA-induced liquid phase condensation of cGAS activates innate immune signaling. Science 361, 704–709 (2018).
Tao, J., Zhang, X. W. & Jin, J. Nonspecific DNA binding of cGAS N terminus promotes cGAS activation. J. Immunol. 198, 3627–3636 (2017).
Lama, L., Adura, C. & Xie, W. Development of human cGAS-specific small-molecule inhibitors for repression of dsDNA-triggered interferon expression. Nat. Commun. 10, 2261 (2019).
Fang, Q. et al. Human cytomegalovirus IE1 protein alters the higher-order chromatin structure by targeting the acidic patch of the nucleosome. Elife 5, e11911 (2016).
Yang, D. et al. Nα-acetylated Sir3 stabilizes the conformation of a nucleosome-binding loop in the BAH domain. Nat. Struct. Mol. Biol. 20, 1116–1118 (2013).
Barbera, A. J. et al. The nucleosomal surface as a docking station for Kaposi’s sarcoma herpesvirus LANA. Science 311, 856–861 (2006).
McGinty, R. K., Henrici, R. C. & Tan, S. Crystal structure of the PRC1 ubiquitylation module bound to the nucleosome. Nature 514, 591–596 (2014).
Makde, R. D., England, J. R., Yennawar, H. P. & Tan, S. Structure of RCC1 chromatin factor bound to the nucleosome core particle. Nature 467, 562–566 (2010).
Kato, H. et al. A conserved mechanism for centromeric nucleosome recognition by centromere protein CENP-C. Science 340, 1110–1113 (2013).
Mackenzie, K. J. et al. cGAS surveillance of micronuclei links genome instability to innate immunity. Nature 548, 461–465 (2017).
Pathare, G. R. & Decout, A. Structural mechanism of cGAS inhibition by the nucleosome. Nature https://doi.org/10.1038/s41586-020-2750-6 (2020).
Zhao, B. & Xu, P. The molecular basis of tight nuclear tethering and inactivation of cGAS. Nature https://doi.org/10.1038/s41586-020-2749-z (2020).
Michalski, S., de Oliveira Mann, C. C., Stafford, C. & Witte, G. Structural basis for sequestration and autoinhibition of cGAS by chromatin. Nature https://doi.org/10.1038/s41586-020-2748-0 (2020).
Kujirai, T. & Zierhut, C. Structural basis for the inhibition of cGAS by nucleosomes. Science https://doi.org/10.1126/science.abd0237 (2020).
Boyer, J. A. & Spangler, C. J. Structural basis of nucleosome-dependent cGAS inhibition. Science https://doi.org/10.1126/science.abd0609 (2020).
Liu, Z. S. et al. G3BP1 promotes DNA binding and activation of cGAS. Nat. Immunol. 20, 18–28 (2019).
Hu, S. & Sun, H. PKR-dependent cytosolic cGAS foci are necessary for intracellular DNA sensing. Sci. Signal. 12, eaav7934 (2019).
Dyer, P. N. et al. Reconstitution of nucleosome core particles from recombinant histones and DNA. Methods Enzymol. 375, 23–44 (2004).
Lowary, P. T. & Widom, J. New DNA sequence rules for high affinity binding to histone octamer and sequence-directed nucleosome positioning. J. Mol. Biol. 276, 19–42 (1998).
Wu, C., Huang, X., Cheng, J., Zhu, D. & Zhang, X. High-quality, high-throughput cryo-electron microscopy data collection via beam tilt and astigmatism-free beam-image shift. J. Struct. Biol. 208, 107396 (2019).
Zheng, S. Q. et al. MotionCor2: anisotropic correction of beam-induced motion for improved cryo-electron microscopy. Nat. Methods 14, 331–332 (2017).
Rohou, A. & Grigorieff, N. CTFFIND4: fast and accurate defocus estimation from electron micrographs. J Struct. Biol. 192, 216–221 (2015).
Scheres, S. H. RELION: implementation of a Bayesian approach to cryo-EM structure determination. J Struct. Biol. 180, 519–530 (2012).
Wakamori, M. et al. Intra- and inter-nucleosomal interactions of the histone H4 tail revealed with a human nucleosome core particle with genetically-incorporated H4 tetra-acetylation. Sci. Rep. 5, 17204 (2015).
Kucukelbir, A., Sigworth, F. J. & Tagare, H. D. Quantifying the local resolution of cryo-EM density maps. Nat. Methods 11, 63–65 (2014).
Pettersen, E. F. et al. UCSF Chimera-a visualization system for exploratory research and analysis. J. Comput. Chem. 25, 1605–1612 (2004).
Emsley, P., Lohkamp, B., Scott, W. G. & Cowtan, K. Features and development of Coot. Acta Crystallogr. D Biol. Crystallogr. 66, 486–501 (2010).
Adams, P. D. et al. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. D Biol. Crystallogr. 66, 213–221 (2010).
Acknowledgements
We thank X. Huang, B. Zhu, X. Li, L. Chen and other staff members at the Center for Biological Imaging (CBI), Core Facilities for protein Science at the Institute of Biophysics, Chinese Academy of Science (IBP, CAS) for the support in cryo-EM data collection; L. Kong for cryo-EM data storage and backup. The project was funded by the National Key R&D Program of China (2017YFA0504700, 2019YFA0508900), the Ministry of Science and Technology of China, the National Natural Science Foundation of China (31930069, 31521002 and 31991162), the Strategic Priority Research Program of the Chinese Academy of Sciences (XDB37040101, XDB37010101). X.Z. received scholarships from the ‘National Thousand (Young) Talents Program’ from the Office of Global Experts Recruitment in China. D.C. is sponsored by the Youth Innovation Promotion Association at the Chinese Academy of Sciences (2018124).
Author information
Authors and Affiliations
Contributions
X.Z. and R.-M.X. conceived the project; X.F. expressed and purified the cGAS proteins. X.H. purified the nucleosomes, prepared the samples of the cGAS-nucleosome complexes and carried out the EMSA experiments. D.C. and X.F. performed the cryo-EM sample preparation, sample screening, data collection and data processing. D.C. carried out the pull-down assays, the activity assays, model building and structure refinement. X.Z., R.-M.X. and D.C. analyzed the structures and wrote the manuscript with the help of all the other authors.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Rights and permissions
About this article
Cite this article
Cao, D., Han, X., Fan, X. et al. Structural basis for nucleosome-mediated inhibition of cGAS activity. Cell Res 30, 1088–1097 (2020). https://doi.org/10.1038/s41422-020-00422-4
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41422-020-00422-4
This article is cited by
-
DNA sensing and repair systems unexpectedly team up against cancer
Nature (2024)
-
MRE11 liberates cGAS from nucleosome sequestration during tumorigenesis
Nature (2024)
-
cGAMP-activated cGAS–STING signaling: its bacterial origins and evolutionary adaptation by metazoans
Nature Structural & Molecular Biology (2023)
-
Mitochondrial damage and activation of the cytosolic DNA sensor cGAS–STING pathway lead to cardiac pyroptosis and hypertrophy in diabetic cardiomyopathy mice
Cell Death Discovery (2022)
-
CENP-N promotes the compaction of centromeric chromatin
Nature Structural & Molecular Biology (2022)