Homotype-Targeted Biogenic Nanoparticles to Kill Multidrug-Resistant Cancer Cells
Abstract
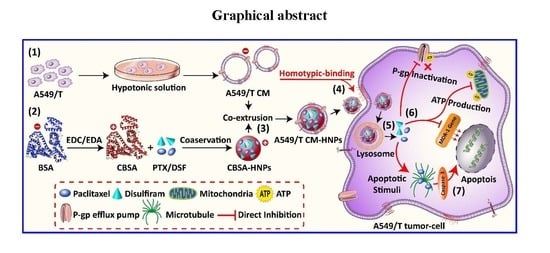
:1. Introduction
2. Materials and Methods
2.1. Materials, Cell Culture, and Animals
2.2. Methods
2.2.1. Synthesis and Characterization of Cationic BSA
2.2.2. Isolation of Cell Plasma Membranes
2.2.3. Cell Membrane Vesicle Preparation and Characterization
2.2.4. Preparation of Nanoparticle Cores
2.2.5. Functionalization of Nanoparticle Cores
2.2.6. Determination of Membrane-Associated Protein
2.2.7. Particle Size, Zeta Potential, and TEM Examination
2.2.8. In Vitro Drug Release Kinetics
2.2.9. Membrane Coating Optimization and Stability Studies
2.2.10. In Vitro Antitumor Assay
2.2.11. Homotypic Uptake Study
2.2.12. Apoptosis and Caspase Assay
2.2.13. P-gp Drug Efflux Inhibition and Intracellular ATP Determination
2.2.14. Hemotoxicity Assay
2.2.15. Statistical Analysis
3. Results and Discussion
3.1. Preparation and Characterization of Cell Membrane-Coated Hybrid Biogenic Nanoparticles
3.2. Membrane Protein Translocation and In Vitro Drug Release Kinetics
3.3. Homotypic Uptake Study
3.4. In Vitro Antitumor Effects
3.5. P-gp Drug Efflux Inhibition and Intracellular ATP Determination
3.6. Hemotoxicity Assay
4. Conclusions
Author Contributions
Funding
Conflicts of Interest
References
- Al-Akra, L.; Bae, D.-H.; Leck, L.Y.; Richardson, D.R.; Jansson, P.J. The biochemical and molecular mechanisms involved in the role of tumor micro-environment stress in development of drug resistance. Biochim. Biophys. Acta (BBA) Gen. Subj. 2019, 1863, 1390–1397. [Google Scholar] [CrossRef] [PubMed]
- Aldinucci, D.; Celegato, M.; Casagrande, N. Microenvironmental interactions in classical Hodgkin lymphoma and their role in promoting tumor growth, immune escape and drug resistance. Cancer Lett. 2016, 380, 243–252. [Google Scholar] [CrossRef] [PubMed]
- Assanhou, A.G.; Li, W.; Zhang, L.; Xue, L.; Kong, L.; Sun, H.; Mo, R.; Zhang, C. Reversal of multidrug resistance by co-delivery of paclitaxel and lonidamine using a TPGS and hyaluronic acid dual-functionalized liposome for cancer treatment. Biomaterials 2015, 73, 284–295. [Google Scholar] [CrossRef] [PubMed]
- Beatty, G.L.; Gladney, W.L. Immune escape mechanisms as a guide for cancer immunotherapy. Clin. Cancer Res. 2014, 21, 687–692. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Beckmann, J.; Schubert, R.; Chiquet-Ehrismann, R.; Müller, D.J. Deciphering Teneurin Domains that Facilitate Cellular Recognition, Cell–Cell Adhesion, and Neurite Outgrowth Using Atomic Force Microscopy-Based Single-Cell Force Spectroscopy. Nano Lett. 2013, 13, 2937–2946. [Google Scholar] [CrossRef] [PubMed]
- Boonstra, M.C.; De Geus, S.W.; Prevoo, H.A.; Hawinkels, L.; Van De Velde, C.J.; Kuppen, P.J.; Vahrmeijer, A.L.; Sier, C.F.M.; Van De Velde, C.J. Selecting Targets for Tumor Imaging: An Overview of Cancer-Associated Membrane Proteins. Biomark. Cancer 2016, 8, 119–133. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chen, Z.; Zhao, P.; Luo, Z.; Zheng, M.; Tian, H.; Gong, P.; Gao, G.; Pan, H.; Liu, L.; Ma, A.; et al. Cancer Cell Membrane–Biomimetic Nanoparticles for Homologous-Targeting Dual-Modal Imaging and Photothermal Therapy. ACS Nano 2016, 10, 10049–10057. [Google Scholar] [CrossRef]
- Del Turco, S.; Ciofani, G.; Cappello, V.; Parlanti, P.; Gemmi, M.; Caselli, C.; Ragusa, R.; Papa, A.; Battaglia, D.; Sabatino, L.; et al. Effects of cerium oxide nanoparticles on hemostasis: Coagulation, platelets, and vascular endothelial cells. J. Biomed. Mater. Res. Part A 2019, 107, 1551–1562. [Google Scholar] [CrossRef]
- Duan, X.; Xiao, J.; Yin, Q.; Zhang, Z.; Yu, H.; Mao, S.; Li, Y. Smart pH-Sensitive and Temporal-Controlled Polymeric Micelles for Effective Combination Therapy of Doxorubicin and Disulfiram. ACS Nano 2013, 7, 5858–5869. [Google Scholar] [CrossRef]
- El-Readi, M.Z.; Eid, S.; Abdelghany, A.A.; Al-Amoudi, H.S.; Efferth, T.; Wink, M. Resveratrol mediated cancer cell apoptosis, and modulation of multidrug resistance proteins and metabolic enzymes. Phytomedicine 2019, 55, 269–281. [Google Scholar] [CrossRef]
- Fan, Z.; Li, P.Y.; Deng, J.; Bady, S.C.; Cheng, H. Cell membrane coating for reducing nanoparticle-induced inflammatory responses to scaffold constructs. Nano Res. 2018, 11, 5573–5583. [Google Scholar] [CrossRef] [PubMed]
- Fang, R.H.; Hu, C.-M.J.; Luk, B.T.; Gao, W.; Copp, J.A.; Tai, Y.; O’Connor, D.E.; Zhang, L. Cancer Cell Membrane-Coated Nanoparticles for Anticancer Vaccination and Drug Delivery. Nano Lett. 2014, 14, 2181–2188. [Google Scholar] [CrossRef] [PubMed]
- Gopisetty, M.K.; Kovács, D.; Igaz, N.; Rónavári, A.; Bélteky, P.; Rázga, Z.; Venglovecz, V.; Csoboz, B.; Boros, I.M.; Konya, Z.; et al. Endoplasmic reticulum stress: Major player in size-dependent inhibition of P-glycoprotein by silver nanoparticles in multidrug-resistant breast cancer cells. J. Nanobiotechnol. 2019, 17, 9. [Google Scholar] [CrossRef] [PubMed]
- Hu, C.-M.J.; Zhang, L.; Aryal, S.; Cheung, C.; Fang, R.H.; Zhang, L. Erythrocyte membrane-camouflaged polymeric nanoparticles as a biomimetic delivery platform. Proc. Natl. Acad. Sci. USA 2011, 108, 109801–109985. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kang, T.; Zhu, Q.; Wei, D.; Feng, J.; Yao, J.; Jiang, T.; Song, Q.; Wei, X.; Chen, H.; Gao, X.; et al. Nanoparticles Coated with Neutrophil Membranes Can Effectively Treat Cancer Metastasis. ACS Nano 2017, 11, 1397–1411. [Google Scholar] [CrossRef] [PubMed]
- Li, T.; Dong, H.; Zhang, C.; Mo, R. Cell-based drug delivery systems for biomedical applications. Nano Res. 2018, 11, 5240–5257. [Google Scholar] [CrossRef]
- Loo, T.W.; Bartlett, M.C.; Clarke, D.M. Disulfiram Metabolites Permanently Inactivate the Human Multidrug Resistance P-Glycoprotein†. Mol. Pharm. 2004, 1, 426–433. [Google Scholar] [CrossRef] [PubMed]
- Misra, R.; Acharya, S.; Sahoo, S.K. Cancer nanotechnology: Application of nanotechnology in cancer therapy. Drug Discov. Today 2010, 15, 842–850. [Google Scholar] [CrossRef] [PubMed]
- Mohammad, I.S.; He, W.; Yin, L. A Smart Paclitaxel-Disulfiram Nanococrystals for Efficient MDR Reversal and Enhanced Apoptosis. Pharm. Res. 2018, 35, 77. [Google Scholar] [CrossRef] [PubMed]
- Mohammad, I.S.; He, W.; Yin, L. Understanding of human ATP binding cassette superfamily and novel multidrug resistance modulators to overcome MDR. Biomed. Pharmacother. 2018, 100, 335–348. [Google Scholar] [CrossRef]
- Mohammad, I.S.; Hu, H.; Yin, L.; He, W. Drug nanocrystals: Fabrication methods and promising therapeutic applications. Int. J. Pharm. 2019, 562, 187–202. [Google Scholar] [CrossRef]
- Mohammad, I.S.; Naveed, M.; Ijaz, S.; Shumzaid, M.; Hassan, S.; Muhammad, K.S.; Rasool, F.; Akhtar, N.; Ishaq, H.M.; Khan, H.M.S. Phytocosmeceutical formulation development, characterization and its in-vivo investigations. Biomed. Pharmacother. 2018, 107, 806–817. [Google Scholar] [CrossRef] [PubMed]
- Mohammad, I.S.; Teng, C.; Chaurasiya, B.; Yin, L.; Wu, C.; He, W. Drug-delivering-drug approach-based codelivery of paclitaxel and disulfiram for treating multidrug-resistant cancer. Int. J. Pharm. 2019, 557, 304–313. [Google Scholar] [CrossRef] [PubMed]
- Que, X.; Su, J.; Guo, P.; Kamal, Z.; Xu, E.; Liu, S.; Chen, J.; Qiu, M. Study on preparation, characterization and multidrug resistance reversal of red blood cell membrane-camouflaged tetrandrine-loaded PLGA nanoparticles. Drug Deliv. 2019, 26, 199–207. [Google Scholar] [CrossRef] [Green Version]
- Rao, L.; He, Z.; Meng, Q.-F.; Zhou, Z.; Bu, L.-L.; Guo, S.; Liu, W.; Zhao, X.-Z. Effective cancer targeting and imaging using macrophage membrane-camouflaged upconversion nanoparticles. J. Biomed. Mater. Res. Part A 2016, 105, 521–530. [Google Scholar] [CrossRef] [PubMed]
- Reichert, J.M.; Wenger, J.B. Development trends for new cancer therapeutics and vaccines. Drug Discov. Today 2008, 13, 30–37. [Google Scholar] [CrossRef]
- Reshma, P.; Unnikrishnan, B.; Preethi, G.; Syama, H.; Archana, M.; Remya, K.; Shiji, R.; Sreekutty, J.; Sreelekha, T. Overcoming drug-resistance in lung cancer cells by paclitaxel loaded galactoxyloglucan nanoparticles. Int. J. Boil. Macromol. 2019, 136, 266–274. [Google Scholar] [CrossRef]
- Rosenblum, D.; Joshi, N.; Tao, W.; Karp, J.M.; Peer, D. Progress and challenges towards targeted delivery of cancer therapeutics. Nat. Commun. 2018, 9, 1410. [Google Scholar] [CrossRef] [Green Version]
- Roundhill, E.A.; Jabri, S.; Burchill, S.A. ABCG1 and Pgp identify drug resistant, self-renewing osteosarcoma cells. Cancer Lett. 2019, 453, 142–157. [Google Scholar] [CrossRef]
- Wang, H.; Gao, Z.; Liu, X.; Agarwal, P.; Zhao, S.; Conroy, D.W.; Ji, G.; Yu, J.; Jaroniec, C.P.; Liu, Z.; et al. Targeted production of reactive oxygen species in mitochondria to overcome cancer drug resistance. Nat. Commun. 2018, 9, 562. [Google Scholar] [CrossRef]
- Zhang, L.; Yang, X.; Lv, Y.; Xin, X.; Qin, C.; Han, X.; Yang, L.; He, W.; Yin, L. Cytosolic co-delivery of miRNA-34a and docetaxel with core-shell nanocarriers via caveolae-mediated pathway for the treatment of metastatic breast cancer. Sci. Rep. 2017, 7, 46186. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, P.; Liu, G.; Chen, X.S. Nanobiotechnology: Cell membrane-based delivery systems. Nano Today 2017, 13, 7–9. [Google Scholar] [CrossRef] [Green Version]
- Zhen, X.; Cheng, P.; Pu, K. Recent Advances in Cell Membrane-Camouflaged Nanoparticles for Cancer Phototherapy. Small 2018, 15, 1804105. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhong, X.; Xiong, M.; Meng, X.; Gong, R. Comparison of the multi-drug resistant human hepatocellular carcinoma cell line Bel-7402/ADM model established by three methods. J. Exp. Clin. Cancer Res. 2010, 29, 115. [Google Scholar] [CrossRef] [PubMed] [Green Version]













| PTX | DSF | |||
|---|---|---|---|---|
| Formulations | DL | EE | DL | EE |
| HNPs | 23.1 ± 1.1 | 65.4 ± 1.6 | 4.9 ± 1.5 | 84.3 ± 3.7 |
| RBCs CM-HNPs | 26.7 ± 1.2 | 71.2 ± 0.4 | 6.2 ± 0.6 | 86.7 ± 1.8 |
| LO2 CM-HNPs | 19.6 ± 2.9 | 63.6 ± 2.1 | 3.4 ± 0.4 | 79.8 ± 3.2 |
| 4T1 CM-HNPs | 18.4 ± 0.7 | 66.8 ± 3.6 | 3.8 ± 1.9 | 72.6 ± 2.4 |
| A549/T CM-HNPs | 24.8 ± 3.5 | 69.4 ± 3.1 | 5.4 ± 1.2 | 71.4 ± 3.6 |
| Formulation | IC50 a (µg) | RRI b | |
|---|---|---|---|
| A549@24 h | A549/T@24 h | 24 h | |
| HNPs | 2.5 ± 0.7 | 3.0 ± 0.5 | 1.2 |
| RBCs CM-HNPs | 1.5 ± 0.3 | 2.3 ± 0.1 | 1.5 |
| LO2 CM-HNPs | 2.3 ± 0.6 | 3.1 ± 0.8 | 1.3 |
| 4T1 CM-HNPs | 1.8 ± 0.7 | 1.7 ± 0.4 | 0.9 |
| A549/T CM-HNPs | 1.4 ± 0.6 * | 1.2 ± 0.1 ** | 0.8 |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Shair Mohammad, I.; Chaurasiya, B.; Yang, X.; Lin, C.; Rong, H.; He, W. Homotype-Targeted Biogenic Nanoparticles to Kill Multidrug-Resistant Cancer Cells. Pharmaceutics 2020, 12, 950. https://doi.org/10.3390/pharmaceutics12100950
Shair Mohammad I, Chaurasiya B, Yang X, Lin C, Rong H, He W. Homotype-Targeted Biogenic Nanoparticles to Kill Multidrug-Resistant Cancer Cells. Pharmaceutics. 2020; 12(10):950. https://doi.org/10.3390/pharmaceutics12100950
Chicago/Turabian StyleShair Mohammad, Imran, Birendra Chaurasiya, Xuan Yang, Chuchu Lin, Hehui Rong, and Wei He. 2020. "Homotype-Targeted Biogenic Nanoparticles to Kill Multidrug-Resistant Cancer Cells" Pharmaceutics 12, no. 10: 950. https://doi.org/10.3390/pharmaceutics12100950