Abstract
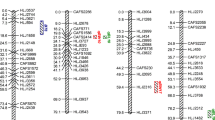
Growth-related traits are economically important in aquaculture, and their variation is linked to quantitative loci (QTLs). For exploring genetic basis of genetic breeding and improvement of grass carp (Ctenopharyngodon idellus), QTL mapping for growth-related traits was performed with an F2 family of grass carp from the Yangtze River system, including 360 offspring. In this study, we mapped QTLs for body weight, body length, body height, and body width using 99 previously published microsatellite loci. A total of 15 growth-related QTLs were found in 7 linkage groups (LG1, LG2, LG14, LG15, LG16, LG18, and LG21). Four QTLs were related to body weight (qBWH1, qBWH14, qBWH15, and qBWH16), and explained 3.1 to 7.7% of the phenotypic variance. Six QTLs were related to body length (qBL1, qBL2, qBL14, qBL15, qBL16, and qBL18), and these QTLs explained 2.8 to 8.9% of the phenotypic variance. Three QTLs affecting body height (qBH14, qBH15, and qBH16) explained 3.2 to 9.2% of the phenotypic variance. Two QTLs affecting body width (qBW15 and qBW21) accounted for 9.2% and 3.3% of the phenotypic variance, respectively. In order to fine map the QTL affecting body weight on LG1 (qBWH1), 14 additional novel microsatellites were included, and the qBWH1 was relocated with a narrower flanking marker interval of 7.5 cM. This is the first study of QTL mapping in grass carp and provides new insights for the application of molecular marker–assisted breeding in this species.


Similar content being viewed by others
References
Alfaqih MA, Brunelli JP, Drew RE, Thorgaard GH (2009) Mapping of five candidate sex-determining loci in rainbow trout (Oncorhynchus mykiss). BMC Genet 10:2. https://doi.org/10.1186/1471-2156-10-2
Beckmann JS, Soller M (1988) Detection of linkage between marker loci and loci affecting quantitative traits in crosses between segregating populations. Theor Appl Genet 76:228–236
Chilton EW, Muoneke MI (1992) Biology and management of grass carp (Ctenopharyngodon idella, Cyprinidae) for vegetation control: a north American perspective. Rev Fish Biol Fish 2:283–320
Derayat A, Houston RD, Guy DR, Hamilton A, Ralph J, Spreckley N, Taggart JB, McAndrew BJ, Haley CS (2007) Mapping QTL affecting body lipid percentage in Atlantic salmon (Salmo salar). Aquaculture. 272:S250–S251. https://doi.org/10.1016/j.aquaculture.2007.07.046
Feng X, Yu X, Fu B, Wang X, Liu H, Pang M, Tong J (2018) A high-resolution genetic linkage map and QTL fine mapping for growth-related traits and sex in the Yangtze River common carp (Cyprinus carpio haematopterus). BMC Genomics 19:230. https://doi.org/10.1186/s12864-018-4613-1
Franch R, Louro B, Tsalavouta M, Chatziplis D (2006) A genetic linkage map of the hermaphrodite teleost fish Sparus aurata L. Genetics 174:851–861
Frankham R (1996) Introduction to quantitative genetics, 4th edn. Trends Genet 12:280. https://doi.org/10.1016/0168-9525(96)81458-2
Jang SH, Liu H, Su JG, Dong F, Xiong F, Liao L, Wang Y, Zhu Z (2010) Construction and characterization of two bacterial artificial chromosome libraries of grass carp. Mar Biotechnol 12:261–266
Kocher TD, Lee WJ, Sobolewska H, Penman D (1998) A genetic linkage map of a cichlid fish, the tilapia (Oreochromis niloticus). Genetics 148:1225–1232
Laghari MY, Zhang Y, Lashari P, Zhang X, Sun X (2013) Quantitative trait loci (QTL) associated with growth rate trait in common carp (Cyprinus carpio). Aquac Int 21:1373–1379
Li D, Shen YB, Fu JJ, Li JL (2013) Isolation and characterization of 25 novel polymorphic microsatellite markers from grass carp (Ctenopharyngodon idella). Conserv Genet Resour 5:745–748
Li JL, Zhu ZY, Wang GL, Bai ZY, Yue GH (2007) Isolation and characterization of 17 polymorphic microsatellites in grass carp. Mol Ecol Resour 7:1114–1116
Li P, Xiao S, Wei N, Zhang Z, Huang R, Gu Y, Guo Y, Ren J, Huang L, Chen C (2012) Fine mapping of a QTL for ear size on porcine chromosome 5 and identification of high mobility group AT-hook 2 (HMGA2) as a positional candidate gene. Genet Sel Evol 44:6. https://doi.org/10.1186/1297-9686-44-6
Liao XL, Ma HY, Xu GB, Shao CW, Chen SL (2009) Construction of a genetic linkage map and mapping of a female-specific DNA marker in half-smooth tongue sole (Cynoglossus semilaevis). Mar Biotechnol 11:699–709
Liu H, Fu B, Pang M, Feng X, Wang X, Yu X, Tong J (2016) QTL fine mapping and identification of candidate genes for growth-related traits in bighead carp (Hypophthalmichehys nobilis). Aquaculture 465:134–143. https://doi.org/10.1016/j.aquaculture.2016.08.039
Lombard V, Delourme R (2001) A consensus linkage map for rapeseed (Brassica napus L.): construction and integration of three individual maps from DH populations. Theor Appl Genet 103:491–507
Lorieux M, Goffinet B, Perrier X, Lanaud C et al (1995) Maximum-likelihood models for mapping genetic markers showing segregation distortion. 1. Backcross populations. Theor Appl Genet 90:73–80
Lu H, Romero-Severson J, Bernardo R (2002) Chromosomal regions associated with segregation distortion in maize. Theor Appl Genet 105:622–628
Ozaki A, Okamoto H, Yamada T, Matuyama T, Sakai T, Fuji K, Sakamoto T, Okamoto N, Yoshida K, Hatori K, Araki K, Okauchi M (2010) Linkage analysis of resistance to Streptococcus iniae infection in Japanese flounder (Paralichthys olivaceus). Aquaculture. 308:S62–S67. https://doi.org/10.1016/j.aquaculture.2010.07.039
Peng W, Xu J, Zhang Y, Feng J, Dong C, Jiang L, Feng J, Chen B, Gong Y, Chen L, Xu P (2016) An ultra-high density linkage map and QTL mapping for sex and growth-related traits of common carp (Cyprinus carpio). Sci Rep 6. https://doi.org/10.1038/srep26693
Ruan XH, Wang WJ, Kong J, Yu F, Huang XQ (2010) Genetic linkage mapping of turbot (Scophthalmus maximus L.) using microsatellite markers and its application in QTL analysis. Aquaculture 308:89–100
Sambrook J, Russell DW (2001) Molecular cloning: a laboratory manual. Cold Spring Harbor, New York
Sánchez-Molano E, Cerna A, Toro MA, Bouza C, Hermida M, Pardo BG, Cabaleiro S, Fernández J, Martínez P (2011) Detection of growth-related QTL in turbot (Scophthalmus maximus). BMC Genomics 12. https://doi.org/10.1186/1471-2164-12-473
Shen Y, Wang L, Fu J, Xu X, Yue GH, Li J (2019) Population structure, demographic history and local adaptation of the grass carp. BMC Genomics 20:467
Smith C, Smith DB (1993) The need for close linkages in marker-assisted selection for economic merit in livestock. Anim Breed Abstr 61:197–204
Song WT, Li YZ, Zhao YW et al (2012) Construction of a high-density microsatellite genetic linkage map and mapping of sexual and growth-related traits in half-smooth tongue sole (Cynoglossus semilaevis). Plos One 7. https://doi.org/10.1371/journal.pone.0052097
Sun J, Shen Y, Fu J, Yu H, Zhang M et al (2015) The effects of the morphometric traits at different month ages on body weight of Ctenopharyngodon idellus. J Shanghai Ocean Univ 24:341–349
van Ooijen JW (2006) Software for the calculation of genetic linkage maps in experimental populations. Wageningen, Netherlands
van Ooijen JW, Boer MP, Jansen RC (2002) MapQTL 4.0: software for the calculation of QTL positions on genetic maps. Wageningen, Netherlands
Wang CM, Lo LC, Yue GH (2007) Identification and verification of QTL associated with growth traits in two genetic backgrounds of barramundi (Lates calcarifer). Anim Genet 39:34–39
Wang CM, Lo LC, Zhu ZY, Yue GH (2006) A genome scan for quantitative trait loci affecting growth-related traits in an F1 family of Asian seabass (Lates calcarifer). BMC Genomics 7:274–270
Wang CM, Bai ZY, He XP, Lin G, Xia JH, Sun F, Lo LC, Feng F, Zhu ZY, Yue GH (2011) A high-resolution linkage map for comparative genome analysis and QTL fine mapping in Asian seabass, Lates calcarifer. BMC Genomics 12:174
Wang Q, Cheng L, Liu J, Li Z, Xie S, de Silva SS (2015a) Freshwater aquaculture in PR China: trends and prospects. Rev Aquac 7:283–302
Wang Y, Lu Y, Zhang Y, Ning Z, Li Y, Zhao Q, Lu H, Huang R, Xia X, Feng Q, Liang X, Liu K, Zhang L, Lu T, Huang T, Fan D, Weng Q, Zhu C, Lu Y, Li W, Wen Z, Zhou C, Tian Q, Kang X, Shi M, Zhang W, Jang S, du F, He S, Liao L, Li Y, Gui B, He H, Ning Z, Yang C, He L, Luo L, Yang R, Luo Q, Liu X, Li S, Huang W, Xiao L, Lin H, Han B, Zhu Z (2015b) The draft genome of the grass carp (Ctenopharyngodon idellus) provides insights into its evolution and vegetarian adaptation. Nat Genet 47:625–631. https://doi.org/10.1038/ng.3280
Wringe BF, Devlin RH, Ferguson MM, Moghadam HK, Danzmann RG (2010) Growth-related quantitative trait loci in domestic and wild rainbow trout (Oncorhynchus mykiss). BMC Genet 11:63. https://doi.org/10.1186/1471-2156-11-63
Xia JH, Liu F, Zhu ZY, Fu JJ, Feng JB, Li JL, Yue GH (2010) A consensus linkage map of the grass carp (Ctenopharyngodon idella) based on microsatellites and SNPs. BMC Genomics 11:135–130
Yue GH (2014) Recent advances of genome mapping and marker-assisted selection in aquaculture. Fish Fish 15:376–396
Zheng XH, Kuang YY, Lv WH, Cao DC (2013) A consensus linkage map of common carp (Cyprinus carpioL.) to compare the distribution and variation of QTLs associated with growth traits. Sci China 56:351–359
Acknowledgments
We thank Mr. Ye Baoqing and Santosh for English editing.
Funding
This work was supported by the Project of Shanghai Engineering and Technology Center for Promoting Ability (19DZ2284300) and China Agriculture Research System (CARS-45-03).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Ethical approval
All applicable international, national, and/or institutional guidelines for the care and use of animals were followed by the authors.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Figure S1
The frequency histogram of the phenotypic data including body weight (BWH), body length (BL), body height (BH), and body width (BW). (PNG 3468 kb)
Figure S2
QTL mapping for growth-related traits. (a), QTL for body weight on LG1, LG14, LG15, LG16; (b), QTL for body length on LG1, LG2, LG14, LG15, LG16, LG18; (c), QTL for body height on LG16, LG18; (d), QTL for body width on LG15, LG21. The Y-axis presents LOD scores and the X-axis presents genetic distances. The red lines were drawn by plotting the LOD scores at each marker along the linkage group. (PNG 6377 kb)
Figure S3
QTL mapping for weight-related trait on LG1. The Y-axis presents LOD scores and the X-axis presents genetic distances. The red line was drawn by plotting the LOD scores at each marker along the linkage group. (PNG 1342 kb)
ESM 4
Details of the linkage map. The asterisk after microsatellites indicates the marker showing segregation distortion. (XLSX 28 kb)
Table S1
Characteristics of the 14 additional novel microsatellites from grass carp (Ctenopharyngodon idellus). (DOC 53 kb)
Rights and permissions
About this article
Cite this article
Yu, C., Xu, X., Li, J. et al. Primary mapping of QTL for growth-related traits in grass carp (Ctenopharyngodon idellus). Aquacult Int 28, 2275–2285 (2020). https://doi.org/10.1007/s10499-020-00594-1
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10499-020-00594-1