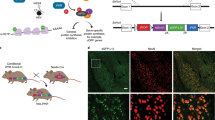
Abstract
To survive in a dynamic environment, animals need to identify and appropriately respond to stimuli that signal danger1. Survival also depends on suppressing the threat-response during a stimulus that predicts the absence of threat (safety)2,3,4,5. An understanding of the biological substrates of emotional memories during a task in which animals learn to flexibly execute defensive responses to a threat-predictive cue and a safety cue is critical for developing treatments for memory disorders such as post-traumatic stress disorder5. The centrolateral amygdala is an important node in the neuronal circuit that mediates defensive responses6,7,8,9, and a key brain area for processing and storing threat memories. Here we applied intersectional chemogenetic strategies to inhibitory neurons in the centrolateral amygdala of mice to block cell-type-specific translation programs that are sensitive to depletion of eukaryotic initiation factor 4E (eIF4E) and phosphorylation of eukaryotic initiation factor 2α (p-eIF2α). We show that de novo translation in somatostatin-expressing inhibitory neurons in the centrolateral amygdala is necessary for the long-term storage of conditioned-threat responses, whereas de novo translation in protein kinase Cδ-expressing inhibitory neurons in the centrolateral amygdala is necessary for the inhibition of a conditioned response to a safety cue. Our results provide insight into the role of de novo protein synthesis in distinct inhibitory neuron populations in the centrolateral amygdala during the consolidation of long-term memories.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
References
Fendt, M. & Fanselow, M. S. The neuroanatomical and neurochemical basis of conditioned fear. Neurosci. Biobehav. Rev. 23, 743–760 (1999).
Pavlov, I. P. Conditioned Reflexes: an Investigation of the Physiological Activity of the Cerebral Cortex (Oxford Univ. Press, 1927)
Rescorla, R. A. Pavlovian conditioned inhibition. Psychol. Bull. 72, 77–94 (1969).
Christianson, J. P. et al. Inhibition of fear by learned safety signals: a mini-symposium review. J. Neurosci. 32, 14118–14124 (2012).
Jovanovic, T. et al. Impaired fear inhibition is a biomarker of PTSD but not depression. Depress. Anxiety 27, 244–251 (2010).
Wilensky, A. E., Schafe, G. E., Kristensen, M. P. & LeDoux, J. E. Rethinking the fear circuit: the central nucleus of the amygdala is required for the acquisition, consolidation, and expression of Pavlovian fear conditioning. J. Neurosci. 26, 12387–12396 (2006).
Ciocchi, S. et al. Encoding of conditioned fear in central amygdala inhibitory circuits. Nature 468, 277–282 (2010).
Han, S., Soleiman, M. T., Soden, M. E., Zweifel, L. S. & Palmiter, R. D. Elucidating an affective pain circuit that creates a threat memory. Cell 162, 363–374 (2015).
Haubensak, W. et al. Genetic dissection of an amygdala microcircuit that gates conditioned fear. Nature 468, 270–276 (2010).
Shrestha, P. et al. Cell-type-specific drug-inducible protein synthesis inhibition demonstrates that memory consolidation requires rapid neuronal translation. Nat. Neurosci. 23, 281–292 (2020).
Kandel, E. R., Dudai, Y. & Mayford, M. R. The molecular and systems biology of memory. Cell 157, 163–186 (2014).
Klann, E. & Dever, T. E. Biochemical mechanisms for translational regulation in synaptic plasticity. Nat. Rev. Neurosci. 5, 931–942 (2004).
Costa-Mattioli, M. et al. eIF2α phosphorylation bidirectionally regulates the switch from short- to long-term synaptic plasticity and memory. Cell 129, 195–206 (2007).
Kats, I. R. & Klann, E. Translating from cancer to the brain: regulation of protein synthesis by eIF4F. Learn. Mem. 26, 332–342 (2019).
Sidrauski, C., McGeachy, A. M., Ingolia, N. T. & Walter, P. The small molecule ISRIB reverses the effects of eIF2α phosphorylation on translation and stress granule assembly. eLife 4, e05033 (2015).
Thoreen, C. C. et al. A unifying model for mTORC1-mediated regulation of mRNA translation. Nature 485, 109–113 (2012).
Li, H. et al. Experience-dependent modification of a central amygdala fear circuit. Nat. Neurosci. 16, 332–339 (2013).
Fadok, J. P. et al. A competitive inhibitory circuit for selection of active and passive fear responses. Nature 542, 96–100 (2017).
Yu, K., Garcia da Silva, P., Albeanu, D. F. & Li, B. Central amygdala somatostatin neurons gate passive and active defensive behaviors. J. Neurosci. 36, 6488–6496 (2016).
Lin, C.-J. et al. Targeting synthetic lethal interactions between Myc and the eIF4F complex impedes tumorigenesis. Cell Rep. 1, 325–333 (2012).
Dickins, R. A. et al. Probing tumor phenotypes using stable and regulated synthetic microRNA precursors. Nat. Genet. 37, 1289–1295 (2005).
Gorkiewicz, T., Balcerzyk, M., Kaczmarek, L. & Knapska, E. Matrix metalloproteinase 9 (MMP-9) is indispensable for long term potentiation in the central and basal but not in the lateral nucleus of the amygdala. Front. Cell. Neurosci. 9, 73 (2015).
Botta, P. et al. Regulating anxiety with extrasynaptic inhibition. Nat. Neurosci. 18, 1493–1500 (2015).
Guettier, J.-M. et al. A chemical-genetic approach to study G protein regulation of beta cell function in vivo. Proc. Natl Acad. Sci. USA 106, 19197–19202 (2009).
Costa-Mattioli, M. et al. Translational control of hippocampal synaptic plasticity and memory by the eIF2α kinase GCN2. Nature 436, 1166–1173 (2005).
Zhu, P. J. et al. Suppression of PKR promotes network excitability and enhanced cognition by interferon-γ-mediated disinhibition. Cell 147, 1384–1396 (2011).
Banko, J. L. et al. Behavioral alterations in mice lacking the translation repressor 4E-BP2. Neurobiol. Learn. Mem. 87, 248–256 (2007).
Hoeffer, C. A. et al. Inhibition of the interactions between eukaryotic initiation factors 4E and 4G impairs long-term associative memory consolidation but not reconsolidation. Proc. Natl Acad. Sci. USA 108, 3383–3388 (2011).
Sharma, V. et al. eIF2α controls memory consolidation via excitatory and somatostatin neurons. Nature https://doi.org/10.1038/s41586-020-2805-8 (2020).
Laxmi, T. R., Stork, O. & Pape, H.-C. Generalization of conditioned fear and its behavioral expression in mice. Behav. Brain Res. 145, 89–98 (2003).
Ghosh, S. & Chattarji, S. Neuronal encoding of the switch from specific to generalized fear. Nat. Neurosci. 18, 112–120 (2015).
Acknowledgements
We thank A. Nnenna Chime and S. Taveras for technical assistance; D. Anderson for PKCδ::GluClα-iCre BAC transgenic mice; H. Zeng for the pAAV.CAG Pr.DIO.tTA plasmid; and N. Heintz, A. Nectow and E. Schmidt for the pAAV.Eef1a1 Pr.DIO.eGFP-L10a plasmid. We are grateful to all members of the Klann laboratory for feedback and discussions; and J. Ledoux and R. Del Triano for feedback on this manuscript. This study was supported by National Institute of Health grants NS034007 and NS047384 to E.K., Canadian Institute of Health Research FDN-148366 to J.P., NARSAD Young Investigator grant 26696 to P.S. and a BP-ENDURE fellowship to K.S.A.R. N.H. is supported by a Howard Hughes Medical Investigator grant.
Author information
Authors and Affiliations
Contributions
P.S. and E.K. conceptualized the framework of this study. P.S. carried out surgeries and behavioural testing, and collected and analysed data. Z.S. carried out western blots and behaviour testing. M.M. performed mouse breeding and pharmacological treatments. K.S.A.R., A.T.Z., C.-Y.J. and P.M.H.-V. carried out mouse behavioural testing. J.P. generated and provided the floxed Col1a1TRE GFP.shmiR-4E mice. N.H. generated and provided the floxed iPKR mice. P.S. and E.K. wrote the paper. All authors read and commented on the paper.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Additional information
Peer review information Nature thanks Richard Palmiter and the other, anonymous, reviewer(s) for their contribution to the peer review of this work.
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data figures and tables
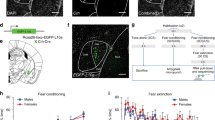
Extended Data Fig. 1 Differential cued threat conditioning.
a, Schematic of the behaviour protocol for the Unpaired group (left) and Box-Only control group (right). b, Freezing response to CS+ and CS- in individual animals trained using the Unpaired behaviour protocol. c, Freezing response to CS+ and CS- in individual animals trained using the Paired behaviour protocol. d, Paired group learned the association between CS+ and US and showed increasing freezing response to successive CS presentations whereas the Unpaired group did not associate CS+ with US. RM Two-way ANOVA with Bonferroni’s post hoc test. Effect of CS+ training: F(1,11) = 11.40, P = 0.0062; effect of CS- training: F(2,33) = 9.360, P = 0.0006. n[Unpaired] = 5 and n[Paired] = 8 animals. e, Both Paired and Unpaired groups, but not Box-Only group, increased freezing levels during the post-tone period compared to the pre-tone period. Two way ANOVA with Bonferroni’s post hoc test. Effect of training: F(2,30) = 13.86, P < 0.0001, effect of epoch: F(1,30) = 60.38, P < 0.0001. n[Box-Only] = 5, n[Unpaired] = 5 and n[Paired] = 8 animals. f, Representative motion traces for Box-Only, Unpaired and Paired groups during LTM. g, Freezing response during pre-CS of LTM test is low for all three groups. One-way ANOVA. P = 0.874. n[Box-Only] = 5, n[Unpaired] = 5 and n[Paired] = 8 animals. h, Animals in the Paired group freeze significantly higher during CS- than during the pre-tone period. Two-way ANOVA with Bonferroni’s post hoc test. Effect of training: F(2,30) = 8.38, P = 0.0013; effect of epoch: F(1,30) = 23.97, P < 0.0001. n[Box-Only] = 5, n[Unpaired] = 5 and n[Paired] = 8 animals. i, Freezing response to CS+ and CS- in individual animals trained using the Paired 5X behaviour protocol. j, Increasing the number of CS-US pairs from 3 to 5 pairings during training led to a continued escalation of freezing response to successive presentations of CS’s. RM Two-way ANOVA with Bonferroni’s post hoc test. Effect of CS+: F(1,24) = 23.95, P < 0.0001; effect of CS-: F(1,24) = 42.74, P < 0.0001. Paired 3X CS+: CS1 vs CSn, P = 0.039; Paired 5X CS+: CS1 vs CSn, P = 0.0005. n[Paired 3X] = 8 and n[Paired 5X] = 6 animals. k, Paired 5X group displayed equivalent conditioned threat response and safety response to CS+ and CS- respectively as paired 3X group during LTM test. Two way ANOVA with Bonferroni’s post hoc test. Effect of pairings: F(1,24) = 0.2942, P = 0.593; effect of CS: F(1,24) = 66.46, P < 0.0001. n[Paired 3X] = 8 and n[Paired 5X] = 6 animals. l, Discrimination index for cued threat in Paired 5X group was unaltered compared to Paired 3X group. Unpaired t-test, Two-tailed. P > 0.999. Data are presented as mean ± s.e.m. *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001. n.s. nonsignificant.
Extended Data Fig. 2 Distinct IN subpopulations in centrolateral amygdala.
a, Co-immunostaining for GFP and neuronal marker, NeuN, in PKCδ.TRAP amygdala sections. 57.96 ± 2.86% of all neurons in centrolateral amygdala are PKCδ INs. n = 3 animals/group. b, Co-immunostaining for tdTomato and NeuN in SOM.tdT amygdala sections. SOM INs constitute 55.36 ± 0.91% of all neurons in CeL. n = 3 animals/group. c, Immunohistochemistry for PKCδ in SOM.tdT brain sections shows largely non-overlapping expression of PKCδ in SOM Cre expressing cells in CeL. d, Immunohistochemistry for SOM in PKCδ.tdT brain sections also shows largely non-overlapping populations but the subcellular distribution of SOM in neuronal processes makes it difficult to analyse the extent of SOM co-expression in PKCδ Cre expressing cell populations. e, Multiplexed smFISH for Prkcd and Som showing mutually exclusive INs in CeL expressing these two mRNA populations. f, Immunohistochemistry data for PKCδ.TRAP amygdala sections showing expression of p-S6 (S235/6) in PKCδ neurons in CeL across three groups (Box-Only, Unpaired and Paired) at 30 min post training. One-way ANOVA with Bonferroni’s post hoc test. F(2,334) = 71.67, P < 0.0001. n[Box-Only] = 117, n[Unpaired] = 118 and n[Paired] = 102 cells from 3 animals/group. g, Immunohistochemistry data for SOM tdTomato sections showing p-S6 (S235/6) in SOM neurons in CeL across groups. One-way ANOVA with Bonferroni’s post hoc test. F(2,292) = 44.18, P < 0.0001. n[Box-Only] = 162, n[Unpaired] = 158 and n[Paired] = 165 cells from 3 animals/group. Scale bar, 50 μm.
Extended Data Fig. 3 Differential threat conditioning induces de novo translation in CeL neurons.
a, Schematic for the in vivo de novo translation labelling assay with puromycin infusion in central amygdala. b, De novo translation was upregulated in PKCδ INs in the Paired training group compared to Box-Only and Unpaired controls. Insets show higher magnification.
Extended Data Fig. 4 Cell-type-specific knockdown of cap dependent translation in CeL neurons.
a, Proportion of endogenous SOM.tdT INs chemogenetically targeted to express shmir-eIF4E in a cre- and tet-dependent manner. 44.75 ± 8.78% of SOM.tdT INs in CeL expressed shmir-eIF4E. n = 3 animals/group. b, Proportion of endogenous PKCδ.tdT INs chemogenetically targeted to express shmir-eIF4E in a cre- and tet-dependent manner. 52.42+4.41% of PKCδ.tdT INs in CeL expressed shmir-eIF4E. n = 3 animals/group. c, eIF4E level was significantly reduced in SOM INs in SOM.4Ekd group compared to SOM.GFP control. Unpaired t-test, Two-tailed. P < 0.0001. n[SOM.GFP] = 87 and n[SOM.4Ekd] = 132 cells from 3 animals/group. d, eIF4E level was significantly knocked down in PKCδ INs in PKCδ.4Ekd group compared to PKCδ.GFP control. Unpaired t-test, Two-tailed. P = 0.0056, n[PKCδ.GFP] = 121 and n[PKCδ.4Ekd] = 87 cells from 3 animals/group. e, Global de novo translation, as measured with puromycin assay, was significantly reduced in SOM.4Ekd group compared to control. Unpaired t-test, Two-tailed. P = 0.0363. n[SOM.GFP] = 53 and n[SOM.4Ekd] = 20 cells from 3 animals/ group. f, Similarly, global de novo protein synthesis was significantly diminished in PKCδ.4Ekd group compared to control. Unpaired t-test, Two-tailed. P < 0.0001. n[PKCδ.GFP] = 120 and n[PKCδ.4Ekd] = 20 cells from 4 animals/ group. g, MMP9 levels was significantly reduced in SOM.4Ekd mice compared to control. Unpaired t-test, Two-tailed. P < 0.0001. n[SOM.GFP] = 87 and n[SOM.4Ekd] = 60 cells from 3 animals/group. h, Similarly, MMP9 level was significantly reduced in PKCδ.4Ekd group compared to control. Unpaired t-test, Two-tailed. P < 0.0001. n[PKCδ.GFP] = 60 and n[PKCδ.4Ekd] = 30 cells from 3 animals/group. Data are presented as mean + s.e.m. *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001. n.s. nonsignificant. Scale bar, 50 μm.
Extended Data Fig. 5 Inhibition of cap-dependent translation and anxiety related behaviours.
a, Representative open field activity traces for SOM.GFP and SOM.4Ekd animals. b, Distance travelled in the open field arena for individual SOM.GFP and SOM.4Ekd animals. c, XY plot showing normal acclimation of SOM.GFP and SOM.4Ekd animals to the open field arena. Effect of Time: F(2,46) = 45.50, P < 0.0001. n[SOM.GFP] = 13 and n[SOM.4Ekd] = 12 animals. d, SOM.GFP and SOM.4Ekd animals display equivalent spontaneous locomotion in the open field arena. Unpaired t-test, Two-tailed. P = 0.895. n[SOM.GFP] = 13 and n[SOM.4Ekd] = 12 animals. e, SOM.4Ekd mice display normal thigmotaxis behaviour compared to control. Unpaired t-test, Two-tailed. P = 0.521. n[SOM.GFP] = 13 and n[SOM.4Ekd] = 12 animals. f, Representative activity heat map in elevated plus maze for SOM.GFP and SOM.4Ekd animals. g, SOM.GFP and SOM.4Ekd animals spend similar duration in the open arm, as a percent of total duration. Unpaired t-test, Two-tailed. P = 0.288. n[SOM.GFP] = 18 and n[SOM.4Ekd] = 18 animals. h, SOM.GFP and SOM.4Ekd mice make equivalent entries into the open arm. Unpaired t-test, Two-tailed. P = 0.107. n[SOM.GFP] = 18 and n[SOM.4Ekd] = 18 animals. i, Representative open field activity traces for PKCδ.GFP and PKCδ.4Ekd animals. j, Distance travelled in the open field arena for individual PKCδ.GFP and PKCδ.4Ekd animals. k, XY plot showing normal acclimation of PKCδ.GFP and PKCδ.4Ekd animals to the open field arena. RM Two-way ANOVA. Time: F(2,32) = 19.12, P < 0.0001. n[PKCδ.GFP] = 10 and n[PKCδ.4Ekd] = 8 animals. l, Bar plot showing total distance travelled by PKCδ WT and PKCδ 4Ekd mice in the open field arena. Unpaired t-test, Two-tailed. P = 0.772. n[PKCδ.GFP] = 10 and n[PKCδ.4Ekd] = 8 animals. m, PKCδ.4Ekd mice show normal thigmotaxis in the open field arena compared to PKCδ.GFP control. Unpaired t-test, Two-tailed. P = 0.888. n[PKCδ.GFP] = 7 and n[PKCδ.4Ekd] = 9 animals. n, Representative activity heat maps in elevated plus maze for PKCδ.GFP and PKCδ.4Ekd animals. o, Bar plot showing significantly increased %time spent in the open arm for PKCδ.4Ekd animals compared to PKCδ.GFP controls. Unpaired t-test, Two-tailed. P = 0.0074. n[PKCδ.GFP] = 9 and n[PKCδ.4Ekd] = 6 animals. p, Bar plot showing % entries into the open arm for PKCδ.4Ekd animals compared to PKCδ.GFP controls. P = 0.0476. n[PKCδ.GFP] = 9 and n[PKCδ.4Ekd] = 6 animals. Data are presented as mean + s.e.m. **P < 0.01, ****P < 0.0001, n.s. nonsignificant.
Extended Data Fig. 6 Inhibition of cap-dependent translation in CeL INs and simple threat conditioning.
a, Schematic for simple threat conditioning paradigm in SOM and PKCδ 4Ekd mice. b, Normal memory acquisition in simple threat-conditioning in WT, SOM.4Ekd and PKCδ 4Ekd groups. Effect of CS: F(2,50) = 32.28, P < 0.0001. n[WT] = 12, n[SOM.4Ekd] = 11 and n[PKCδ.4Ekd] = 5 animals. c, Representative motion traces for WT, SOM.4Ekd and PKCδ.4Ekd groups during LTM test. d, Freezing response to CS+ and CS- in individual SOM.GFP animals during training. e, Freezing response to CS+ and CS- in individual SOM.4Ekd animals during training. f, Normal memory acquisition in differential threat conditioning in SOM.GFP and SOM.4Ekd mice. Effect of CS+: F(2,26) = 34.66, P < 0.0001; effect of CS-: F(2,26) = 20.81, P < 0.0001. n[SOM.GFP] = 10 and n[SOM.4Ekd] = 5 animals. g, Freezing response to CS+ and CS- in individual PKCδ.GFP animals during training. h, Freezing response to CS+ and CS- in individual PKCδ.4Ekd animals during training. i, Normal memory acquisition in PKCδ.GFP and PKCδ.4Ekd mice. Effect of CS+: F(2,34) = 24.67, P < 0.0001; effect of CS-: F(2,34) = 36.84, P < 0.0001. n[PKCδ.GFP] = 9 and n[PKCδ.4Ekd] = 10 animals. j, SOM.4Ekd mice have negligible freezing response during pre-CS in Training phase compared to controls. Unpaired t-test, Two-tailed. P = 0.341. n[SOM.GFP] = 11 and n[SOM.4Ekd] = 10 animals. k, PKCδ.4Ekd mice have negligible freezing response during pre-CS in the Training phase compared to controls. Unpaired t-test, Two-tailed. P = 0.541. n[PKCδ.GFP] = 8 and n[PKCδ.4Ekd] = 11 animals. l, SOM.4Ekd mice have comparable low freezing response during pre-CS in LTM test compared to controls. Unpaired t-test, Two-tailed. P = 0.389. n[SOM.GFP] = 13 and n[SOM.4Ekd] = 12 animals. m, PKCδ.4Ekd mice have comparable low freezing response during pre-CS in LTM test compared to controls. Unpaired t-test, Two-tailed. P = 0.068. n[PKCδ.GFP] = 9 and n[PKCδ.4Ekd] = 11 animals. Data are presented as mean + s.e.m. **P < 0.01, ****P < 0.0001, n.s. nonsignificant.
Extended Data Fig. 7 Cell type-specific eIF2α phosphorylation and threat conditioning.
a, Compared to vehicle controls, ASV infusion in the central amygdala of SOM.iPKR.TRAP animals significantly increased phosphorylation of eIF2α in SOM neurons. Unpaired t-test, Two-tailed. P = 0.0013. n[SOM.iPKR.TRAP +VEH] = 43 and n[SOM.iPKR.TRAP +ASV] = 53 cells from 3 animals/ group. b, ASV infusion in CeA of PKCδ.iPKR.TRAP mice also significantly elevated p-eIF2α in PKCδ neurons compared to vehicle control. Unpaired t-test, Two-tailed. P < 0.0001. n[PKCδ.iPKR.TRAP +VEH] = 36 and n[PKCδ.iPKR.TRAP +ASV] = 38 cells from 3 animals/ group. c, Freezing response to CS+ and CS- in individual SOM.WT animals during training. d, Freezing response to CS+ and CS- in individual SOM.iPKR animals during training. e, Normal memory acquisition in SOM.WT and SOM.iPKR animals in differential threat conditioning paradigm. RM Two-way ANOVA with Bonferroni’s post hoc test. Effect of CS+: F(2,26) = 10.98, P = 0.0003; effect of CS-: F(2,26) = 18.40, P < 0.0001. n[SOM.WT] = 5 and n[SOM.iPKR] = 10 animals. f, Freezing response to CS+ and CS- in individual PKCδ.WT animals during training. g, Freezing response to CS+ and CS- in individual PKCδ.iPKR animals during training. h, Normal memory acquisition in PKC.WT and PKC.iPKR animals in differential threat conditioning paradigm. RM Two-way ANOVA with Bonferroni’s post hoc test. Effect of CS+: F(2,30) = 18.70, P < 0.0001; effect of CS-: F(2,30) = 46.39, P < 0.0001. n[PKC.WT] = 7 and n[PKC.iPKR] = 10 animals. Data are presented as mean + s.e.m. **P < 0.01, ****P < 0.0001. Scale bar, 50 μm.
Extended Data Fig. 8 Chemogenetic modulation of G-protein signalling in CeL SOM INs affects associative learning.
a, Freezing response to CS+ and CS- in individual SOM.tdT animals treated with vehicle during training. b, Freezing response to CS+ and CS- in individual SOM.tdT animals treated with C21 during training. c, C21 treated SOM.tdT mice learn normally compared to VEH treated controls. RM Two-way ANOVA with Bonferroni’s post hoc test. Effect of CS+: F(2,22) = 8.02, P = 0.0024; effect of CS-: F(2,22) = 17.00, P < 0.0001. n[SOM.tdT +VEH] = 7 and n[SOM.tdT +C21] = 6 animals. d, Freezing response to CS+ and CS- in individual SOM.hM4Di animals treated with vehicle during training. e, Freezing response to CS+ and CS- in individual SOM.hM4Di animals treated with C21 during training. f, C21 treated SOM.hM4Di mice have normal memory acquisition relative to VEH controls. RM Two-way ANOVA with Bonferroni’s post hoc test. CS+: F(2,22) = 20.62, P < 0.0001; CS-: F(2,22) = 19.62, P < 0.0001. n[SOM.hM4Di +VEH] = 6 and n[SOM.hM4Di +C21] = 7 animals. g, Freezing response to CS+ and CS- in individual SOM.hM3Dq animals treated with vehicle during training. h, Freezing response to CS+ and CS- in individual SOM.hM3Dq animals treated with C21 during training. i, C21 treated SOM.hM3Dq animals acquire differential threat memory normally relative to VEH controls. RM Two-way ANOVA with Bonferroni’s post hoc test. Effect of CS+: F(2,20) = 17.09, P < 0.0001; effect of CS-: F(2,20) = 38.94, P < 0.0001. n[SOM.hM4Di +VEH] = 5 and n[SOM.hM4Di +C21] = 7 animals. j, C21 treated SOM.tdT mice exhibit normal threat and safety LTM response to CS+ and CS- respectively. RM Two-way ANOVA with Bonferroni’s post hoc test. Effect of drug: F(1,22) = 5.233, P = 0.0321; effect of CS: F(1,22) = 52.87, P < 0.0001. n[SOM.tdT +VEH] = 7 and n[SOM.tdT +C21] = 6 animals. k, C21 treatment does not alter cued threat discrimination index in SOM.tdT mice. Unpaired t-test, Two-tailed. P = 0.6313. n[SOM.tdT +VEH] = 7 and n[SOM.tdT +C21] = 6 animals. l, Freezing response during pre-CS of training session is negligible across all C21 and VEH treated SOM groups. Two-way ANOVA. Effect of drug: F(2,31) = 2.410, P = 0.1064. n[SOM.tdT +VEH] = 7, n[SOM.tdT +C21] = 6, n[SOM.hM4Di +VEH] = 6, n[SOM.hM4Di +C21] = 7, n[SOM.hM3Dq +VEH] = 4 and n[SOM.hM3Dq +C21] = 7 animals. m, C21 treated SOM.tdT, SOM.hM4Di and SOM.hM3Dq mice have equivalent freezing response during pre-CS of LTM test compared to VEH controls. Two-way ANOVA. Effect of drug: F(2.32) = 1.899, P = 0.1663. n[SOM.tdT +VEH] = 7, n[SOM.tdT +C21] = 6, n[SOM.hM4Di +VEH] = 6, n[SOM.hM4Di +C21] = 8, n[SOM.hM3Dq +VEH] = 5 and n[SOM.hM3Dq +C21] = 6 animals. Data are presented as mean ± s.e.m. *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001. n.s. nonsignificant.
Extended Data Fig. 9 Chemogenetic modulation of G-protein signalling in CeL PKCδ INs affects associative learning.
a, Freezing response to CS+ and CS- in individual PKCδ.tdT animals treated with vehicle during training. b, Freezing response to CS+ and CS- in individual PKCδ.tdT animals treated with C21 during training. c, C21 treated PKCδ.tdT animals have normal memory acquisition relative to VEH controls, with progressive increase in freezing response to successive presentation of CS’s. RM Two-way ANOVA with Bonferroni’s post hoc test. Effect of CS+: F(2,20) = 12.22, P = 0.0003; effect of CS-: F(2,20) = 18.65, P < 0.0001. n[PKCδ.tdT +VEH] = 7 and n[PKCδ.tdT +C21] = 5 animals. d, Freezing response to CS+ and CS- in individual PKCδ.hM4Di animals treated with vehicle during training. e, Freezing response to CS+ and CS- in individual PKCδ.hM4Di animals treated with C21 during training. f, C21 treated PKCδ.hM4Di animals learn normally compared to VEH controls. RM Two-way ANOVA with Bonferroni’s post hoc test. Effect of CS+: F(2,24) = 29.92, P < 0.0001; effect of CS-: F(2,24) = 19.58, P < 0.0001. n[PKCδ.hM4Di +VEH] = 8 and n[PKCδ.hM4Di +C21] = 6 animals. g, Freezing response to CS+ and CS- in individual PKCδ.hM3Dq animals treated with vehicle during training. h, Freezing response to CS+ and CS- in individual PKCδ.hM3Dq animals treated with C21 during training. I, C21 treated PKCδ.hM3Dq animals acquire differential threat memory normally compared to VEH controls. RM Two-way ANOVA with Bonferroni’s post hoc test. Effect of CS+: F(2,20) = 15.90, P < 0.0001; effect of CS-: F(2,20) = 20.67, P < 0.0001. n[PKCδ.hM3Dq +VEH] = 5 and n[PKCδ.hM3Dq +C21] = 7 animals. j, C21 treated PKCδ.tdT mice exhibit normal threat and safety LTM response to CS+ and CS- respectively. RM Two-way ANOVA with Bonferroni’s post hoc test. Effect of CS: F(1,20) = 0.402. n[PKCδ.tdT +VEH] = 7 and n[PKCδ.tdT +C21] = 5 animals. k, C21 treatment does not alter cued threat discrimination index in PKCδ.tdT mice. Unpaired t-test, Two-tailed. P = 0.3116. n[PKCδ.tdT +VEH] = 7 and n[PKCδ.tdT +C21] = 6 animals. l, Freezing response during pre-CS of the training session is negligible across all C21 and VEH treated PKCδ groups. Two-way ANOVA. Effect of drug: F(2,35) = 0.2326, P = 0.794. n[PKCδ.tdT +VEH] = 7, n[PKCδ.tdT +C21] = 6, n[PKCδ.hM4Di +VEH] = 8, [PKCδ.hM4Di +C21] = 6, n[PKCδ.hM3Dq +VEH] = 5 and n[PKCδ.hM3Dq +C21] = 7 animals. m, C21 treatment in PKCδ.tdT, PKCδ.hM4Di and PKCδ.hM3Dq animals does not alter baseline freezing response during pre-CS of LTM test. Two-way ANOVA. Effect of drug: F(2,32) = 0.0171, P = 0.983. n[PKCδ.tdT +VEH] = 7, n[PKCδ.tdT +C21] = 5, n[PKCδ.hM4Di +VEH] = 8, [PKCδ.hM4Di +C21] = 6, n[PKCδ.hM3Dq +VEH] = 5 and n[PKCδ.hM3Dq +C21] = 7 animals. Data are presented as mean ± s.e.m. *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001. n.s. nonsignificant.
Extended Data Fig. 10 Working model of simultaneous consolidation and storage of threat and safety cue-associated memories in CeL SOM and PKCδ INs, respectively.
In a differential threat conditioning paradigm, cued threat mobilizes the translation machinery in CeL SOM INs via activation of Gq-coupled GPCR(s), and the ensuing de novo protein synthesis in these neurons is necessary for long-term storage of a cued threat response. The cued safety signal, on the other hand, is processed by CeL PKCδ INs with engagement of the cell-autonomous protein synthesis machinery, leading to long-term storage of the cued safety response.
Supplementary information
Supplementary Figures
This file contains uncropped gels.
41586_2020_2793_MOESM3_ESM.xlsx
Supplementary Table Table 1. Statistical analyses of data presented in Main Figures. Statistical analyses performed using GraphPad Prism 8 for all datasets displayed in main figures are listed. For each data analysis, columns and rows that represent different groups are identified. Data from two groups were compared using two-tailed unpaired Student’s t test, for which p-values are provided. Multiple group comparisons were conducted using one-way ANOVA, or two-way ANOVA, with Bonferroni’s post-hoc test when appropriate. The F statistics for effects of each variable and interaction of variables are listed for multiple group comparisons. Sample size for each group is provided. Post-hoc pairwise comparisons carried out using Bonferroni test are listed as appropriate. Statistical analyses were performed with an α level of 0.05. p values <0.05 were considered significant. *p<0.05, **p<0.01, ***p<0.001, ****p<0.0001. n.s. nonsignificant.
41586_2020_2793_MOESM4_ESM.xlsx
Supplementary Table Table 2. Statistical analyses of data presented in Extended Data Figures. Statistical analyses performed using GraphPad Prism 8 for all datasets displayed in Extended Data figures are listed. For each data analysis, columns and rows that represent different groups are identified. Data from two groups were compared using two-tailed unpaired Student’s t test, for which p-values are provided. Multiple group comparisons were conducted using one-way ANOVA, or two-way ANOVA, with Bonferroni’s post-hoc test when appropriate. The F statistics for effects of each variable and interaction of variables are listed for multiple group comparisons. Sample size for each group is provided. Post-hoc pairwise comparisons carried out using Bonferroni test are listed as appropriate. Statistical analyses were performed with an α level of 0.05. p values <0.05 were considered significant. *p<0.05, **p<0.01, ***p<0.001, ****p<0.0001. n.s. nonsignificant.
Rights and permissions
About this article
Cite this article
Shrestha, P., Shan, Z., Mamcarz, M. et al. Amygdala inhibitory neurons as loci for translation in emotional memories. Nature 586, 407–411 (2020). https://doi.org/10.1038/s41586-020-2793-8
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41586-020-2793-8
This article is cited by
-
Social buffering in rats reduces fear by oxytocin triggering sustained changes in central amygdala neuronal activity
Nature Communications (2024)
-
Cell-type-specific translational control of spatial working memory by the cap-binding protein 4EHP
Molecular Brain (2023)
-
Plastic and stimulus-specific coding of salient events in the central amygdala
Nature (2023)
-
Volume changes of hippocampal and amygdala subfields in patients with mild cognitive impairment and Alzheimer’s disease
Acta Neurologica Belgica (2023)
-
History of suicide attempt associated with amygdala and hippocampus changes among individuals with schizophrenia
European Archives of Psychiatry and Clinical Neuroscience (2023)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.