Abstract
Introduction
Approximately 1% of the world’s population is impacted by epilepsy, a chronic neurological disorder characterized by seizures. One-third of epileptic patients are resistant to AEDs, or have medically refractory epilepsy (MRE). One non-invasive treatment that exists for MRE includes the ketogenic diet, a high-fat, low-carbohydrate diet. Despite the KD’s success in seizure attenuation, it has a few risks and its mechanisms remain poorly understood. The KD has been shown to improve metabolism and mitochondrial function in epileptic phenotypes. Potassium channels have implications in epileptic conditions as they have dual roles as metabolic sensors and control neuronal excitation.
Objectives
The goal of this study was to explore changes in the lipidome in hippocampal and cortical tissue from Kv1.1-KO model of epilepsy.
Methods
FT-ICR/MS analysis was utilized to examine nonpolar metabolome of cortical and hippocampal tissue isolated from a Kv1.1 channel knockout mouse model of epilepsy (n = 5) and wild-type mice (n = 5).
Results
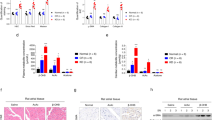
Distinct metabolic profiles were observed, significant (p < 0.05) features in hippocampus often being upregulated (FC ≥ 2) and the cortex being downregulated (FC ≤ 0.5). Pathway enrichment analysis shows lipid biosynthesis was affected. Partition ratio analysis revealed that the ratio of most metabolites tended to be increased in Kv1.1−/−. Metabolites in hippocampal tissue were commonly upregulated, suggesting seizure initiation in the hippocampus. Aberrant mitochondrial function is implicated by the upregulation of cardiolipin, a common component in the mitochondrial membrane.
Conclusion
Generally, our study finds that the lipidome is changed in the hippocampus and cortex in response to Kv1.1-KO indicating changes in membrane structural integrity and synaptic transmission.





Similar content being viewed by others
References
Baek, B. S., Kwon, H. J., Lee, K. H., Yoo, M. A., Kim, K. W., Ikeno, Y., et al. (1999). Regional difference of ROS generation, lipid peroxidaton, and antioxidant enzyme activity in rat brain and their dietary modulation. Archives of Pharmacal Research, 22(4), 361–366. https://doi.org/10.1007/bf02979058.
Barbarosie, M., & Avoli, M. (1997). CA3-driven hippocampal-entorhinal loop controls rather than sustains in vitro limbic seizures. The Journal of Neuroscience, 17(23), 9308–9314. https://doi.org/10.1523/jneurosci.17-23-09308.1997.
Bazan, N. G., Tu, B., & Turco, E. B. R. D. (2002). What synaptic lipid signaling tells us about seizure-induced damage and epileptogenesis. Progress in Brain Research, 135, 175–185. https://doi.org/10.1016/s0079-6123(02)35017-9.
Botion, L. M., & Doretto, M. C. (2003). Changes in peripheral energy metabolism during audiogenic seizures in rats. Physiology & Behavior, 78(4–5), 535–541. https://doi.org/10.1016/s0031-9384(03)00061-1.
Bragin, A., Engel, J., Wilson, C. L., Fried, I., & Mathern, G. W. (1999). Hippocampal and entorhinal cortex high-frequency oscillations (100-500 Hz) in human epileptic brain and in kainic acid-treated rats with chronic seizures. Epilepsia, 40(2), 127–137. https://doi.org/10.1111/j.1528-1157.1999.tb02065.x.
Calzada, E., Avery, E., Sam, P. N., Modak, A., Wang, C., Mccaffery, J. M., et al. (2019). Phosphatidylethanolamine made in the inner mitochondrial membrane is essential for yeast cytochrome bc1 complex function. Nature Communications. https://doi.org/10.1038/s41467-019-09425-1.
Chong, J., Wishart, D. S., & Xia, J. (2019). Using MetaboAnalyst 4.0 for comprehensive and integrative metabolomics data analysis. Current Protocols in Bioinformatics. https://doi.org/10.1002/cpbi.86.
Dodge, J. C., Treleaven, C. M., Fidler, J. A., Tamsett, T. J., Bao, C., Searles, M., et al. (2013). Metabolic signatures of amyotrophic lateral sclerosis reveal insights into disease pathogenesis. Proceedings of the National Academy of Sciences of the USA, 110, 10812–10817. https://doi.org/10.1073/pnas.1308421110.
Ebert, D., Haller, R. G., & Walton, M. E. (2003). Energy contribution of octanoate to intact rat brain metabolism measured by 13C nuclear magnetic resonance spectroscopy. Journal of Neuroscience, 23, 5928–5935.
Farooqui, A. A., Horrocks, L. A., & Farooqui, T. (2000). Glycerophospholipids in brain: their metabolism, incorporation into membranes, functions, and involvement in neurological disorders. Chemistry and Physics of Lipids, 106(1), 1–29. https://doi.org/10.1016/s0009-3084(00)00128-6.
Fenoglio-Simeone, K. A., Wilke, J. C., Milligan, H. L., Allen, C. N., Rho, J. M., & Maganti, R. K. (2009). Ketogenic diet treatment abolishes seizure periodicity and improves diurnal rhythmicity in epilepticKcna1-null mice. Epilepsia, 50(9), 2027–2034. https://doi.org/10.1111/j.1528-1167.2009.02163.x.
Folch, J., Lees, M., & Stanley, G. H. S. (1957). A simple method for the isolation and purification of total lipides from animal tissues. Journal of Biological Chemistry, 226(1), 497–509.
Guzman, P. D., Dantuono, M., & Avoli, M. (2004). Initiation of electrographic seizures by neuronal networks in entorhinal and perirhinal cortices in vitro. Neuroscience, 123(4), 875–886. https://doi.org/10.1016/j.neuroscience.2003.11.013.
Huttenlocher, P. R. (1976). Ketonemia and seizures: Metabolic and anticonvulsant effects of two ketogenic diets in childhood epilepsy. Pediatric Research, 10(5), 536–540. https://doi.org/10.1203/00006450-197605000-00006.
Jernerén, F., Söderquist, M., & Karlsson, O. (2015). Post-sampling release of free fatty acids—Effects of heat stabilization and methods of euthanasia. Journal of Pharmacological and Toxicological Methods, 71, 13–20. https://doi.org/10.1016/j.vascn.2014.11.001.
Ji, J., Kline, A. E., Amoscato, A., Samhan-Arias, A. K., Sparvero, L. J., Tyurin, V. A., et al. (2012). Lipidomics identifies cardiolipin oxidation as a mitochondrial target for redox therapy of brain injury. Nature Neuroscience, 15(10), 1407–1413. https://doi.org/10.1038/nn.3195.
Kalilani, L., Sun, X., Pelgrims, B., Noack-Rink, M., & Villanueva, V. (2018). The epidemiology of drug-resistant epilepsy: A systematic review and meta-analysis. Epilepsia, 59(12), 2179–2193. https://doi.org/10.1111/epi.14596.
Kelly, K. M., & Chung, S. S. (2011). Surgical treatment for refractory epilepsy: Review of patient evaluation and surgical options. Epilepsy Research and Treatment, 2011, 1–10. https://doi.org/10.1155/2011/303624.
Kim, D. Y., Simeone, K. A., Simeone, T. A., Pandya, J. D., Wilke, J. C., Ahn, Y., et al. (2015). Ketone bodies mediate antiseizure effects through mitochondrial permeability transition. Annals of Neurology, 78(1), 77–87. https://doi.org/10.1002/ana.24424.
Kunduri, G., Turner-Evans, D., Konya, Y., Izumi, Y., Nagashima, K., Lockett, S., et al. (2018). Defective cortex glia plasma membrane structure underlies light-induced epilepsy in cpes mutants. Proceedings of the National Academy of Sciences. https://doi.org/10.1073/pnas.1808463115.
Lutas, A., & Yellen, G. (2013). The ketogenic diet: Metabolic influences on brain excitability and epilepsy. Trends in Neurosciences, 36(1), 32–40. https://doi.org/10.1016/j.tins.2012.11.005.
Magrané, J., Cortez, C., Gan, W. B., & Manfredi, G. (2014). Abnormal mitochondrial transport and morphology are common pathological denominators in SOD1 and TDP43 ALS mouse models. Human Molecular Genetics, 23, 1413–1424. https://doi.org/10.1093/hmg/ddt528.
Mcdonald, T., Puchowicz, M., & Borges, K. (2018). Impairments in oxidative glucose metabolism in epilepsy and metabolic treatments thereof. Frontiers in Cellular Neuroscience. https://doi.org/10.3389/fncel.2018.00274.
Miranda, A. M., Bravo, F. V., Chan, R. B., Sousa, N., Paolo, G. D., & Oliveira, T. G. (2019). Differential lipid composition and regulation along the hippocampal longitudinal axis. Translational Psychiatry. https://doi.org/10.1038/s41398-019-0478-6.
Naudí, A., Cabré, R., Jové, M., Ayala, V., Gonzalo, H., Portero-Otín, M., et al. (2015). Lipidomics of human brain aging and Alzheimers disease pathology. International Review of Neurobiology Omic Studies of Neurodegenerative Disease: Part B. https://doi.org/10.1016/bs.irn.2015.05.008.
Nichols, C. G. (2006). KATP channels as molecular sensors of cellular metabolism. Nature, 440(7083), 470–476. https://doi.org/10.1038/nature04711.
Oliver, D., Lien, C., Soom, M., Baukrowitz, T., Jonas, P., & Fakler, B. (2004). Functional conversion between A-type and delayed rectifier K channels by membrane lipids. Science, 304(5668), 265–270. https://doi.org/10.1126/science.1094113.
Oliveros, J. C. (2007). VENNY. An interactive tool for comparing lists with Venn Diagrams. Retrieved from http://bioinfogp.cnb.csic.es/tools/venny/index.html.
Paradies, G., Paradies, V., Benedictis, V. D., Ruggiero, F. M., & Petrosillo, G. (2014). Functional role of cardiolipin in mitochondrial bioenergetics. Biochimica Et Biophysica Acta (BBA) - Bioenergetics, 1837(4), 408–417. https://doi.org/10.1016/j.bbabio.2013.10.006.
Pearson-Smith, J., & Patel, M. (2017). Metabolic dysfunction and oxidative stress in epilepsy. International Journal of Molecular Sciences, 18(11), 2365. https://doi.org/10.3390/ijms18112365.
Pluskal, T., Castillo, S., Villar-Briones, A., & Oresic, M. (2010). MZmine 2: modular framework for processing, visualizing, and analyzing mass spectrometry-based molecular profile data. BMC Bioinform, 11, 395–395. https://doi.org/10.1186/1471-2105-11-395.
Robbins, C. A., & Tempel, B. L. (2012). Kv1.1 and Kv1.2: similar channels, different seizure models. Epilepsia, 53, 134–141. https://doi.org/10.1111/j.1528-1167.2012.03484.x.
Roundtree, H. M., Simeone, T. A., Johnson, C., Matthews, S. A., Samson, K. K., & Simeone, K. A. (2016). Orexin receptor antagonism improves sleep and reduces seizures in Kcna1-null mice. Sleep, 39(2), 357–368. https://doi.org/10.5665/sleep.5444.
Shirai, Y., & Saito, N. (2014). Diacylglycerol kinase as a possible therapeutic target for neuronal diseases. Journal of Biomedical Science, 21(1), 28. https://doi.org/10.1186/1423-0127-21-28.
Simeone, K. A., Matthews, S. A., Samson, K. K., & Simeone, T. A. (2014a). Targeting deficiencies in mitochondrial respiratory complex I and functional uncoupling exerts anti-seizure effects in a genetic model of temporal lobe epilepsy and in a model of acute temporal lobe seizures. Experimental Neurology, 251, 84–90. https://doi.org/10.1016/j.expneurol.2013.11.005.
Simeone, T. A., Samson, K. K., Matthews, S. A., & Simeone, K. A. (2014b). In vivo ketogenic diet treatment attenuates pathologic sharp waves and high frequency oscillations in in vitro hippocampal slices from epileptic Kv1.1α knockout mice. Epilepsia. https://doi.org/10.1111/epi.12603.
Sooderberg, M., Edlund, C., Alafuzoff, I., Kristensson, K., & Dallner, G. (1992). Lipid composition in different regions of the brain in Alzheimers disease/senile dementia of Alzheimers type. Journal of Neurochemistry, 59(5), 1646–1653. https://doi.org/10.1111/j.1471-4159.1992.tb10994.x.
Stas, J. I., Bocksteins, E., Jensen, C. S., Schmitt, N., & Snyders, D. J. (2016). The anticonvulsant retigabine suppresses neuronal KV2-mediated currents. Scientific Reports. https://doi.org/10.1038/srep35080.
Sud, M., Fahy, E., Cotter, D., Brown, A., Dennis, E. A., Glass, C. K., et al. (2007). LMSD: LIPID MAPS structure database. Nucleic Acids Research. https://doi.org/10.1093/nar/gkl838.
Thiam, A. R., Farese, R. V., Jr., & Walther, T. C. (2013). The biophysics and cell biology of lipid droplets. Nature Reviews Molecular Cell Biology, 14, 775–786. https://doi.org/10.1038/nrm3699.
Tracey, T. J., Steyn, F. J., Wolvetang, E. J., & Ngo, S. T. (2018). Neuronal lipid metabolism: multiple pathways driving functional outcomes in health and disease. Frontiers in Molecular Neuroscience, 11(10), 1–25.
Turker, S., Severcan, M., Ilbay, G., & Severcan, F. (2014). Epileptic seizures induce structural and functional alterations on brain tissue membranes. Biochimica Et Biophysica Acta (BBA)—Biomembranes, 1838(12), 3088–3096. https://doi.org/10.1016/j.bbamem.2014.08.025.
Vismer, M. S., Forcelli, P. A., Skopin, M. D., Gale, K., & Koubeissi, M. Z. (2015). The piriform, perirhinal, and entorhinal cortex in seizure generation. Frontiers in Neural Circuits. https://doi.org/10.3389/fncir.2015.00027.
Wasterlain, C. G., Thompson, K. W., Suchomelova, L., & Niquet, J. (2010). Brain energy metabolism during experimental neonatal seizures. Neurochemical Research, 35(12), 2193–2198. https://doi.org/10.1007/s11064-010-0339-4.
Weljie, A. M., Meerlo, P., Goel, N., Sengupta, A., Kayser, M. S., Abel, T., et al. (2015). Oxalic acid and diacylglycerol 36:3 are cross-species markers of sleep debt. Proceedings of the National Academy of Sciences, 112(8), 2569–2574. https://doi.org/10.1073/pnas.1417432112.
Wenk, M. R. (2005). The emerging field of lipidomics. Nature Reviews Drug Discovery, 4(7), 594–610. https://doi.org/10.1038/nrd1776.
Wenzel, H. J., Vacher, H., Clark, E., Trimmer, J. S., Lee, A. L., Sapolsky, R. M., et al. (2007). Structural consequences of Kcna1 gene deletion and transfer in the mouse hippocampus. Epilepsia, 48(11), 2023–2046. https://doi.org/10.1111/j.1528-1167.2007.01189.x.
Wishart, D. S., Feunang, Y. D., Marcu, A., Guo, A. C., Liang, K., Vázquez-Fresno, R., et al. (2017). HMDB 4.0: the human metabolome database for 2018. Nucleic Acids Research. https://doi.org/10.1093/nar/gkx108.
Zhang, M., Mileykovskaya, E., & Dowhan, W. (2005). Cardiolipin is essential for organization of complexes III and IV into a supercomplex in intact yeast mitochondria. Journal of Biological Chemistry, 280(33), 29403–29408. https://doi.org/10.1074/jbc.m504955200.
Acknowledgements
This work was supported by grants from the National Institute of Health (P20 RR017675, P20 GM104320, NS085389, NS072179 and NS111389) and from Citizens United for Research in Epilepsy Foundation.
Author information
Authors and Affiliations
Contributions
A.J., R.A.G., D.M., C.H.T.B., H.K. and K.Sa. carried out experiments; A.J., R.A.G., D.M., C.H.T.B., C.B., K.Si., T.S., T.H., C.K.H. and J.A. performed data analysis and interpretation; A.J. and J.A. wrote paper with input from all the authors.
Corresponding author
Ethics declarations
Conflict of interest
The authors declare no competing financial interests.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Below is the link to the electronic supplementary material.
11306_2020_1729_MOESM2_ESM.docx
Supplementary material 2 (DOCX 24 kb) Supplementary Table 1. The 66 significant (p<0.05) features identified in the hippocampus. A majority were found to be elevated in Kv1.1-/- mice compared to WT mice. *Provided annotation represent simple m/z based fit and can be affected by in source fragmentation. Therefore, it is important to note that all identifications are tentative, and their validation would require additional characterization involving purified standards and extensive MSn analysis
11306_2020_1729_MOESM3_ESM.docx
Supplementary material 3 (DOCX 24 kb) Supplementary Table 2. 72 significant (p<0.05) features identified in the cortex with almost all compounds being downregulated in the Kv1.1 -/- compared to the WT. Retention times and m/z have been included
11306_2020_1729_MOESM4_ESM.docx
Supplementary material 4 (DOCX 12 kb) Supplementary Table 3. Pathways that were significantly (adj. p<0.05) affected via Reactome pathway database search
11306_2020_1729_MOESM5_ESM.xlsx
Supplementary material 5 (XLSX 11 kb) Supplementary Table 4. Statistical Data from multivariate analysis performed using one-way ANOVA analysis.
Rights and permissions
About this article
Cite this article
Johnson, A., Grove, R.A., Madhavan, D. et al. Changes in lipid profiles of epileptic mouse model. Metabolomics 16, 106 (2020). https://doi.org/10.1007/s11306-020-01729-4
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s11306-020-01729-4