Abstract
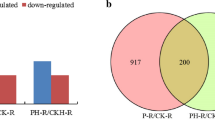
MicroRNAs (miRNAs) have been reported to be correlated with various stress responses in soybean, but only a few miRNAs have been demonstrated to respond to low phosphorus (LP) stress. To unravel the response mechanisms of miRNAs to low-P stress, the roots of two representative soybean genotypes with different P efficiency, Nannong94-156 (a LP-tolerant genotype) and Bogao (a LP-sensitive genotype), were used for the construction of RNA sequencing (RNA-seq) libraries under low/normal-P treatment by high-throughput sequencing. In total, 603 existing miRNAs and 1699 novel miRNAs belonging to 248 and 1582 families in all samples were identified, respectively. Among these miRNAs, 777 miRNAs were differentially expressed (DE) across different P levels and genotypes. Furthermore, putative targets of DE miRNAs were predicted, and these miRNAs mainly targeted ERF (ethylene responsive factor), auxin response factors (ARF), zinc finger protein, MYB, and NAC domain transcription factors. Gene ontology (GO) analysis showed that targets of DE miRNAs were significantly enriched in binding, metabolic processes, biological regulation, response to stress, and phosphorus metabolic processes. In addition, the expression profiles of chosen P-responsive miRNAs and target genes were validated by quantitative real-time PCR (qRT-PCR). Our study focused on genome-wide miRNA identification in two representative soybean genotypes under low-P stress. Overall, the DE miRNAs across different P levels and genotypes and their putative target genes will provide useful information for further study of miRNAs mediating low-P response and facilitate improvements in soybean breeding.









Similar content being viewed by others
Data availability
All relevant data are available in the paper, its Supporting Information files, and from the NCBI BioProject, SRA accession number SRP233151.
Abbreviations
- qRT-PCR:
-
quantitative reverse transcription
- TPM:
-
transcripts per million
- ATPase:
-
ATP synthase
- Rfam:
-
RNA family database
- KEGG:
-
Kyoto encyclopedia of genes and genomes
- GO:
-
gene ontology
- sRNAs:
-
small RNAs
- snoRNAs:
-
small nucleolar RNAs
- snRNAs:
-
small nuclear RNAs
References
Andrews S (2011) FastQC: a quality control tool for high throughput sequence data. http://www.bioinformatics.babraham.ac.uk/projects/fastqc. Accessed 6 October 2011
Ashburner M, Ball CA, Blake JA, Botstein D, Butler H, Cherry JM, Davis AP, Dolinski K, Dwight SS, Eppig JT, Harris MA, Hill DP, Issel-Tarver L, Kasarskis A, Lewis S, Matese JC, Richardson JE, Ringwald M, Rubin GM, Sherlock G, Gene Ontology C (2000) Gene ontology: tool for the unification of biology. Nat Genet 25:25–29
Baek D, Kim MC, Chun HJ, Kang S, Park HC, Shin G, Park J, Shen M, Hong H, Kim WY, Kim DH, Lee SY, Bressan RA, Bohnert HJ, Yun DJ (2013) Regulation of miR399f transcription by AtMYB2 affects phosphate starvation responses in Arabidopsis. Plant Physiol 161:362–373
Bao H, Chen H, Chen M, Xu HM, Huo XW, Xu QH, Wang YW (2019) Transcriptome-wide identification and characterization of microRNAs responsive to phosphate starvation in Populus tomentosa. Funct Integr Genomics 19:953–972
Bolger AM, Lohse M, Usadel B (2014) Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 30:2114–2120
Cakir O, Candar-Cakir B, Zhang BH (2016) Small RNA and degradome sequencing reveals important microRNA function in Astragalus chrysochlorus response to selenium stimuli. Plant Biotechnol J 14:543–556
Calderon-Vazquez C, Sawers RJH, Herrera-Estrella L (2011) Phosphate deprivation in maize: genetics and genomics. Plant Physiol 156:1067–1077
Chiou TJ (2007) The role of microRNAs in sensing nutrient stress. Plant Cell Environ 30:323–332
Fontana JE, Wang G, Sun RR, Xue HY, Li Q, Liu J, Davis KE, Thornburg TE, Zhang BH, Zhang ZY, Pan XP (2020) Impact of potassium deficiency on cotton growth, development and potential microRNA-mediated mechanism. Plant Physiol Biochem 153:72–80
Gao B, Cao C, Li T (2012) Effect of Ethylene on morphology and physiological characteristic of soybean seedlings under low-phosphorus stress. Soybean Science 31:58–63
Gardner PP, Daub J, Tate JG, Nawrocki EP, Kolbe DL, Lindgreen S, Wilkinson AC, Finn RD, Griffiths-Jones S, Eddy SR, Bateman A (2009) Rfam: updates to the RNA families database. Nucleic Acids Res 37:D136–D140
Griffiths-Jones S, Saini HK, van Dongen S, Enright AJ (2008) miRBase: tools for microRNA genomics. Nucleic Acids Res 36:D154–D158
Ha M, Pang MX, Agarwal V, Chen ZJ (2008) Interspecies regulation of microRNAs and their targets. BBA-Gene Regul Mech 1779:735–742
Hackenberg M, Huang PJ, Huang CY, Shi BJ, Gustafson P, Langridge P (2013) A comprehensive expression profile of microRNAs and other classes of non-coding small RNAs in barley under phosphorous-deficient and -sufficient conditions. DNA Res 20:109–125
Hsieh LC, Lin SI, Shih ACC, Chen JW, Lin WY, Tseng CY, Li WH, Chiou TJ (2009) Uncovering small RNA-mediated responses to phosphate deficiency in arabidopsis by deep sequencing. Plant Physiol 151:2120–2132
Hu B, Zhu C, Li F, Tang J, Wang Y, Lin A, Liu L, Che R, Chu C (2011) LEAF TIP NECROSIS1 plays a pivotal role in the regulation of multiple phosphate starvation responses in rice. Plant Physiol 156:1101–1115
Hu B, Wang W, Deng K, Li H, Zhang ZH, Zhang LH, Chu CC (2015) MicroRNA399 is involved in multiple nutrient starvation responses in rice. Front Plant Sci 6:10
Huen A, Bally J, Smith P (2018) Identification and characterisation of microRNAs and their target genes in phosphate-starved Nicotiana benthamiana by small RNA deep sequencing and 5'RACE analysis. BMC Genomics 19:18
Kanehisa M, Goto S (2000) KEGG: Kyoto encyclopedia of genes and genomes. Nucleic Acids Res 28:27–30
Kong XP, Liu GC, Liu JJ, Ding ZJ (2018) The root transition zone: a hot spot for signal crosstalk. Trends Plant Sci 23:403–409
Kuo HF, Chiou TJ (2011) The role of microRNAs in phosphorus deficiency signaling. Plant Physiol 156:1016–1024
Li C, Sha A (2016) Overexpression of GmMIR319 in tobacco improving tolerance to phosphorus deficiency. Chinese Journal of Oil Crop Sciences 38:167–171
Li HY, Hu T, Amombo E, Fu JM (2017a) Genome-wide identification of heat stress-responsive small RNAs in tall fescue (Festuca arundinacea) by high-throughput sequencing. J Plant Physiol 213:157–165
Li SJ, Castillo-Gonzalez C, Yu B, Zhang XR (2017b) The functions of plant small RNAs in development and in stress responses. Plant J 90:654–670
Li ZY, Xu HY, Li Y, Wan XF, Ma Z, Cao J, Li ZS, He F, Wang YF, Wan LQ, Tong ZY, Li XL (2018) Analysis of physiological and miRNA responses to Pi deficiency in alfalfa (Medicago sativa L.). Plant Mol Biol 96:473–492
Lin WY, Lin YY, Chiang SF, Syu CH, Hsieh LC, Chiou TJ (2018) Evolution of microRNA827 targeting in the plant kingdom. New Phytol 217:1712–1725
Liu C, Chu H, Han L, Dai D, Chen H, Tang L (2019) Molecular characterization and target gene prediction of plant miR399 family. Acta Agriculturae Boreali-Sinica 34:1–7
Livak KJ, Schmittgen TD (2001) Analysis of relative gene expression data using real-time quantitative PCR and the 2(T)(-Delta Delta C) method. Methods 25:402–408
Luo J, Liu GY, Chen Z, Ren QY, Yin H, Luo JX, Wang H (2015) Identification and characterization of microRNAs by deep-sequencing in Hyalomma anatolicum anatolicum (Acari: Ixodidae) ticks. Gene 564:125–133
Lyu C, Sha A (2017) Response to phosphorus deficiency regulated by microRNA168 in soybean plant. Chinese Journal of Oil Crop Sciences 39:321–325
Muller M, Munne-Bosch S (2015) Ethylene response factors: a key regulatory hub in hormone and stress signaling. Plant Physiol 169:32–41
Nagarajan VK, Smith AP (2012) Ethylene’s role in phosphate starvation signaling: more than just a root growth regulator. Plant Cell Physiol 53:277–286
Nie Z, Ren ZY, Wang LB, Su SZ, Wei X, Zhang X, Wu L, Liu D, Tang HT, Liu HL, Zhang SZ, Gao SB (2016) Genome-wide identification of microRNAs responding to early stages of phosphate deficiency in maize. Physiol Plant 157:161–174
Ning LH, Du WK, Song HN, Shao HB, Qia WC, Amr Sheteiwy MS, Yu DY (2019) Identification of responsive miRNAs involved in combination stresses of phosphate starvation and salt stress in soybean root. Environ Exp Bot:167
Niu YY, Chen M, Xu ZS, Li LC, Chen XP, Ma YZ (2013) Characterization of ethylene receptors and their interactions with GmTPR-A novel tetratricopeptide repeat protein (TPR) in soybean (Glycine max L.). J Integr Agric 12:571–581
Peng Z, Tian J, Luo RL, Kang YH, Lu YF, Hu YJ, Liu N, Zhang J, Cheng H, Niu SQ, Zhang J, Yao YC (2020) MiR399d and epigenetic modification comodulate anthocyanin accumulation in Malus leaves suffering from phosphorus deficiency. Plant Cell Environ 43:1148–1159
Pokoo R, Ren SC, Wang QY, Motes CM, Hernandez TD, Ahmadi S, Monteros MJ, Zheng Y, Sunkar R (2018) Genotype- and tissue-specific miRNA profiles and their targets in three alfalfa (Medicago sativa L.) genotypes. BMC Genomics 19
Robinson MD, McCarthy DJ, Smyth GK (2010) edgeR: a bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 26:139–140
Romera FJ, Smith AP, Perez-Vicente R (2016) Editorial: Ethylene’s role in plant mineral nutrition. Front Plant Sci 7:2
Sattar S, Addo-Quaye C, Thompson GA (2016) miRNA-mediated auxin signalling repression during Vat-mediated aphid resistance in Cucumis melo. Plant Cell Environ 39:1216–1227
Sha AH, Chen YH, Ba HP, Shan ZH, Zhang XJ, Wu XJ, Qiu DZ, Chen SL, Zhou XN (2012) Identification of Glycine max microRNAs in response to phosphorus deficiency. J Plant Biol 55:268–280
Song HN, Yin ZT, Chao MN, Ning LH, Zhang D, Yu DY (2014) Functional properties and expression quantitative trait loci for phosphate transporter GmPT1 in soybean. Plant Cell Environ 37:462–472
Song L, Yu HP, Dong JS, Che XM, Jiao YL, Liu D (2016) The molecular mechanism of ethylene-mediated root hair development induced by phosphate starvation. PLoS Genet:12
Subramanian S, Fu Y, Sunkar R, Barbazuk WB, Zhu JK, Yu O (2008) Novel and nodulation-regulated microRNAs in soybean roots. BMC Genomics:9
Sun XC, Xu L, Wang Y, Yu RG, Zhu XW, Luo XB, Gong YQ, Wang RG, Limera C, Zhang KY, Liu LW (2015) Identification of novel and salt-responsive miRNAs to explore miRNA-mediated regulatory network of salt stress response in radish (Raphanus sativus L.). BMC Genomics 16
Sunkar R, Li YF, Jagadeeswaran G (2012) Functions of microRNAs in plant stress responses. Trends Plant Sci 17:196–203
Tang HL, Shen JB, Zhang FS, Rengel Z (2013) Interactive effects of phosphorus deficiency and exogenous auxin on root morphological and physiological traits in white lupin (Lupinus albus L.). Sci China-Life Sci 56:313–323
Thirugnanasambantham K, Durairaj S, Saravanan S, Karikalan K, Muralidaran S, Islam VIH (2015) Role of ethylene response transcription factor (ERF) and its regulation in response to stress encountered by plants. Plant Mol Biol Report 33:347–357
Vance CP (2001) Symbiotic nitrogen fixation and phosphorus acquisition. Plant nutrition in a world of declining renewable resources. Plant Physiol 127:390–397
Vance CP, Uhde-Stone C, Allan DL (2003) Phosphorus acquisition and use: critical adaptations by plants for securing a nonrenewable resource. New Phytol 157:423–447
Wang X, Zeng X, Wang W, Luo M, Liao H (2011) Effects of different element deficiencies on soybean nutrient concentration. Journal of South China Agricultural University 32:31–34
Wang QS, Liu N, Yang XY, Tu LL, Zhang XL (2016) Small RNA-mediated responses to low- and high-temperature stresses in cotton. Sci Rep 6:14
Xie ZL, Nolan TM, Jiang H, Yin YH (2019) AP2/ERF transcription factor regulatory networks in hormone and abiotic stress responses in Arabidopsis. Front Plant Sci 10:228
Xu F, Liu Q, Chen LY, Kuang JB, Walk T, Wang JX, Liao H (2013) Genome-wide identification of soybean microRNAs and their targets reveals their organ-specificity and responses to phosphate starvation. BMC Genomics 14:66
Yamaji N, Takemoto Y, Miyaji T, Mitani-Ueno N, Oshida KTY, Ma JF (2017) Reducing phosphorus accumulation in rice grains with an impaired transporter in the node. Nature 541:92-+
Yu HL, Luo N, Sun LC, Liu D (2012) HPS4/SABRE regulates plant responses to phosphate starvation through antagonistic interaction with ethylene signalling. J Exp Bot 63:4527–4538
Yuan H, Liu D (2008) Signaling components involved in plant responses to phosphate starvation. J Integr Plant Biol 50:849–859
Zeng HQ, Wang GP, Zhang YQ, Hu XY, Pi EX, Zhu YY, Wang HZ, Du LQ (2016) Genome-wide identification of phosphate-deficiency-responsive genes in soybean roots by high-throughput sequencing. Plant Soil 398:207–227
Zeng HQ, Zhu YY, Huang SQ, Yang ZM (2010) Analysis of phosphorus-deficient responsive miRNAs and cis-elements from soybean (Glycine max L.). J Plant Physiol 167:1289–1297
Zeng JB, Ye ZL, He XY, Zhang GP (2019) Identification of microRNAs and their targets responding to low-potassium stress in two barley genotypes differing in low-K tolerance. J Plant Physiol 234:44–53
Zhang BH (2015) MicroRNA: a new target for improving plant tolerance to abiotic stress. J Exp Bot 66:1749–1761
Zhang BH, Pan XP, Cobb GP, Anderson TA (2006) Plant microRNA: a small regulatory molecule with big impact. Dev Biol 289:3–16
Zhang D, Zhang HY, Chu SS, Li HY, Chi YJ, Triebwasser-Freese D, Lv HY, Yu DY (2017) Integrating QTL mapping and transcriptomics identifies candidate genes underlying QTLs associated with soybean tolerance to low-phosphorus stress. Plant Mol Biol 93:137–150
Zhang GY, Chen DG, Zhang T, Duan AG, Zhang JG, He CY (2018) Transcriptomic and functional analyses unveil the role of long non-coding RNAs in anthocyanin biosynthesis during sea buckthorn fruit ripening. DNA Res 25:465–476
Zhang HY, Yang YM, Sun CY, Liu XQ, Lv LL, Hu ZB, Yu DY, Zhang D (2020) Up-regulating GmETO1 improves phosphorus uptake and use efficiency by promoting root growth in soybean. Plant Cell Environ 43:2080–2094
Zhao GJ, Yu HY, Liu MM, Lu YG, Ouyang B (2017) Identification of salt-stress responsive microRNAs from Solanum lycopersicum and Solanum pimpinellifolium. Plant Growth Regul 83:129–140
Zhao MJ, Yin LJ, Ma J, Zheng JC, Wang YX, Lan JH, Fu JD, Chen M, Xu ZS, Ma YZ (2019) The roles of GmERF135 in improving salt tolerance and decreasing ABA sensitivity in soybean. Front Plant Sci 10:940
Zhu YY, Zeng HQ, Dong CX, Yin XM, Shen QR, Yang ZM (2010) microRNA expression profiles associated with phosphorus deficiency in white lupin (Lupinus albus L.). Plant Sci 178:23–29
Funding
This work was supported by the key scientific and technological project of Henan Province (192102110023), the Key Scientific Research Projects of Higher Education Institutions in Henan Province (20A210017), the Henan agricultural university science and technology innovation fund (KJCX2019C02), and the Ministry of Science and Technology of China (2016YFD0100500).
Author information
Authors and Affiliations
Contributions
Conceptualization: Dan Zhang; methodology: Xiaoqian Liu; formal analysis and investigation: Xiaoqian Liu; writing—original draft preparation: Xiaoqian Liu; writing—review and editing: Dan Zhang, Shanshan Chu; funding acquisition: Dan Zhang; resources: Jinyu Zhang; supervision: Dan Zhang; Data curation: Dan Zhang, Xiaoqian Liu; software: Shanshan Chu; validation: Chongyuan Sun; investigation: Huanqing Xu; visualization: Yongqing Jiao
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflicts of interest.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary materials
Figure S1.
Functional categorization and distribution of miRNAs targets based on Gene Ontology (GO) classification in Nannong94-156-LP vs Bogao-LP (TIF 146 kb) (PNG 160 kb)
Figure S2.
Bubble chart of KEGG analysis based on targets of DE miRNAs in Nannong94-156-LP vs Bogao-LP (TIF 185 kb) (PNG 193 kb)
ESM 1
(XLSX 43687 kb)
Rights and permissions
About this article
Cite this article
Liu, X., Chu, S., Sun, C. et al. Genome-wide identification of low phosphorus responsive microRNAs in two soybean genotypes by high-throughput sequencing. Funct Integr Genomics 20, 825–838 (2020). https://doi.org/10.1007/s10142-020-00754-9
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10142-020-00754-9