Abstract
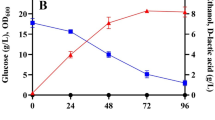
In this study, K. oxytoca KMS004 (ΔadhE Δpta-ackA) was further reengineered by the deletion of frdABCD and pflB genes to divert carbon flux through D-(−)-lactate production. During fermentation of high glucose concentration, the resulted strain named K. oxytoca KIS004 showed poor in growth and glucose consumption due to its insufficient capacity to generate acetyl-CoA for biosynthesis. Evolutionary adaptation was thus employed with the strain to overcome impaired growth and acetate auxotroph. The evolved K. oxytoca KIS004-91T strain exhibited significantly higher glucose-utilizing rate and D-(−)-lactate production as a primary route to regenerate NAD+. D-(−)-lactate at concentration of 133 g/L (1.48 M), with yield and productivity of 0.98 g/g and 2.22 g/L/h, respectively, was obtained by the strain. To the best of our knowledge, this strain provided a relatively high specific productivity of 1.91 g/gCDW/h among those of other previous works. Cassava starch was also used to demonstrate a potential low-cost renewable substrate for D-(−)-lactate production. Production cost of D-(−)-lactate was estimated at $3.72/kg. Therefore, it is possible for the KIS004-91T strain to be an alternative biocatalyst offering a more economically competitive D-(−)-lactate production on an industrial scale.
Key Points
• KIS004-91T produced optically pure D-(−)-lactate up to 1.48 M in a low salts medium.
• It possessed the highest specific D-(−)-lactate productivity than other reported strains.
• Cassava starch as a cheap and renewable substrate was used for D-(−)-lactate production.
• Costs related to media, fermentation, purification, and waste disposal were reduced.




Similar content being viewed by others
References
Abdel-Rahman MA, Tashiro Y, Sonomoto K (2013) Recent advances in lactic acid production by microbial fermentation processes. Biotechnol Adv 31:877–902
Axley MJ, Grahame DA, Stadtman TC (1990) Escherichia coli formate-hydrogen lyase. Purification and properties of the selenium-dependent formate dehydrogenase compoent. J Biol Chem 265:18213–18218
Brisse S, Fevre C, Passet V, Issenhuth-Jeanjean S, Tournebize R, Diancourt L, Grimont P (2009) Virulent clones of Klebsiella pneumoniae: identification and evolutinary scenario based on genomic and phenotypic characterization. PLoS One 4:e4982
Carroll SM, Marx CJ (2013) Evolution after introduction of a novel metabolic pathway consistently leads to restoration of wild-type physiology. PLoS Genet 9:e1003427
Causey TB, Shanmugam KT, Yomano LP, Ingram LO (2004) Engineering Escherichia coli for efficient conversion of glucose to pyruvate. Proc Natl Acad Sci U S A 101:2235–2240
Cecchini G, Schroder I, Gunsalus RP, Maklashina E (2002) Succinate dehydrogenase and fumarate reductase from Escherichia coli. Biochim Biophys Acta 1553:140–157
Celinska E, Grajek W (2009) Biotechnological production of 2,3-butanediol-current state and prospects. Biotechnol Adv 27:715–725
Chan S, Jantama SS, Kanchanatawee S, Jantama K (2016) Process optimization on miro-aeration supply for high production yield of 2,3-butanediol form maltodextrin by metabolically-engineered Klebsiella oxytoca. PLoS One 11:e0161503
Clark DP (1989) The fermentation pathways of Escherichia coli. FEMS Microbiol Lett 63:223–234
de Souza EA, Rossi DM, Ayub MAZ (2014) Bioconversion of residual glycerol from biodisel synthesis into 1,3-propandiol using immobilized cells of Klebsiella pneumoniae BLh-1. Renew Energy 72:253–257
Feng X, Jiang L, Han X, Liu X, Zhao Z, Liu H, Xian M, Zhao G (2017) Production of D-lactate from glucose using Klebsiella pneumoniae mutants. Microb Cell Factories 16:209
Ghaly AE, Kamal M, Correia LR (2005) Kinetic modeling of continuous submerged fermentation of cheese whey for single-cell protein production. Bioresour Technol 96:1143–1152
Jantama K, Zhang X, Moore JC, Shanmugam KT, Svoronos SA, Ingram LO (2008a) Eliminating side products and increasing succinate yields in engineered strains of Escherichia coli C. Biotechnol Bioeng 101:881–893
Jantama K, Haupt MJ, Svoronos SA, Zhang X, Moore JC, Shanmugam KT, Ingram LO (2008b) Combining metabolic engineering and metabolic evolution to develop nonrecombinant strains of Escherichia coli C that produce succinate and malate. Biotechnol Bioeng 99:1140–1153
Jantama K, Polyiam P, Khunnonkwao P, Chan S, Sangproo M, Khor K, Kanchanatawee S (2015) Efficient reduction of the formation of by-products and improvement of production yield of 2,3-butanediol by a combined deletion of alcohol dehydrogenase, acetate kinase-phosphotransacetylase, and lactate dehydrogenase genes in metabolically engineered Klebsiella oxytoca in mineral salts medium. Metab Eng 30:16–26
Khor K, Sawisit A, Chan S, Kanchanatawee S, Jantama SS, Jantama K (2016) High production yield and specific productivity of succinate from cassava starch by metabolically engineered Escherichia coli KJ122. J Chem Technol Biotechnol 91:2834–2841
Khunnonkwao P, Jantama SS, Kanchanatawee S, Jantama K (2018) Re-engineering Escherichia coli KJ122 to enhance the utilization of xylose and xylose/glucose mixture for efficient succinate production in mineral salt medium. Appl Microbiol Biotechnol 102:127–141
Kim Y, Ingram LO, Shanmugam KT (2008) Dihydrolipoamide dehydrogenase mutation alters the NADH sensitivity of pyruvate dehydrogenase complex of Escherichia coli K-12. J Bacteriol 190:3851–3858
Klein-Marcuschamer D, Simmons BA, Blanch HW (2011) Techno-economic analysis of a lignocellulosic ethanol biorefinery with ionic liquid pre-treatment. Biofuels Bioprod Biorefin 5:562–569
Lee CE, Remfert JL, Opgenorth T, Lee KM, Stanford E, Connolly JW, Kim J, Tomke S (2017) Evolutionary respones to crude oil from the Deepwater horizon oil spill by the copepod Eurytemora affinis. Evol Appl 10:813–828
Lu DR, Xiao CM, Xu SJ (2009) Starch-based completely biodegradable polymer materials. Express Polym Lett 6:366–375
Lu H, Zhao X, Wang Y, Ding X, Wang J, Garza E, Manow R, Iverson A, Zhou S (2016) Enhancement of D-lactic acid production from a mixed glucose and xylose substrate by the Escherichia coli strain JH15 devoid of the glucose effect. BMC Biotechnol 16:19
Martinez A, Grabar TB, Shanmugam KT, Yomano LP, York SW, Ingram LO (2007) Low salt medium for lactate and ethanol production by recombinant Escherichia coli B. Biotechnol Lett 29:397–404
Martinez I, Lee A, Bennett GN, San KY (2011) Culture conditions' impact on succinate production by high succinate producing Escherichia coli strain. Biotechnol Prog 27:1225–1231
Mazumdar S, Clomburg JM, Gonzalez R (2010) Escherichia coli strains engineered for homofermentative production of D-lactic acid from glycerol. Appl Environ Microbiol 76:4327–4336
McKinlay JB, Vieille C, Zeikus JG (2007) Prospects for a bio-based succinate industry. Appl Microbiol Biotechnol 76:727–740
Okano K, Yoshida S, Tanaka T, Ogino C, Fukuda H, Kondo A (2009) Homo-D-lactic acid fermentation from arabinose by redirection of the phosphoketolase pathway to the pentose phosphate pathway in L-lactate dehydorgenase gene-deficient Lactobacillus plantraum. Appl Environ Microbiol 75:5175–5178
Okano K, Hama S, Kihara M, Noda H, Tanaka T, Kondo A (2017) Production of optically pure D-lactic acid from brown rice using metabolically engineered Lactobacillus plantarum. Appl Microbiol Biotechnol 101:1869–1875
Park JM, Song H, Lee HJ, Seung D (2013) In silico aided metabolic engineering of Klebsiella oxytoca and fermentation optimization for enhanced 2,3-butanediol production. J Ind Microbiol Biotechnol 40:1057–1066
Park HJ, Bae JH, Ko HJ, Lee SH, Sung BH, Han JI, Sohn JH (2018) Low-pH production of d-lactic acid using newly isolated acid-tolerant yeast Pichia kudriavzevii NG7. Biotechnol Bioeng 115:2232–2242
Qureshi N, Blaschek HP (2001) Recent advances in ABE fermentation: hyper-butanol producing Clostridium beijerinckii BA101. Ind Microbial Biotechnol 27:287–291
Sangproo M, Polyiam P, Jantama SS, Kanchanatawee S, Jantama K (2012) Metabolic engineering of Klebsiella oxytoca M5a1 to produce optically pure D-lactate in mineral salts medium. Bioresour Technol 119:191–198
Sawers G, Bock A (1988) Anaerobic regulation of pyruvate formate-lyase from Escherichia coli K12. J Bacteriol 170:5330–5336
Subramanian MR, Talluri S, Christopher LP (2015) Production of lactic acid using a new homofermentative Enterococcus faecalis isolate. Microb Biotechnol 8:221–229
Van Hellemond JJ, Tielens AG (1994) Expression and functional properties of fumarate reductase. Biochem J 304:321–331
Wood BE, Yomano LP, York SW, Ingram LO (2005) Development of industrial-medium-required elimination of the 2,3-butanediol fermentation pathway to maintain ethanol yield in an ethanologenic strain of Klebsiella oxytoca. Biotechnol Prog 21(5):1366–1372
Yamada R, Wakita K, Mitsui R, Ogino H (2017) Enhanced A-lactic acid production by recombinant Saccharomyces cerevisiae following optimization of the global matabolic pathway. Biotechnol Bioeng 114:2075–2084
Yang YT, Peredelchuk M, Bennet GN, San KY (2000) Effect of variation of Klebsiella pneumoniae acetolactate synthase expression on metabolic flux redistrubution in Escherichia coli. Biotechnol Bioeng 69:150–159
Yang G, Tian J, Li J (2007) Fermentation of 1,3-propanediol by a lactate deficient mutant of Klebsiella oxytoca under microaerobic conditons. Appl Microbiol Biotechnol 73:1017–1024
Zhang X, Wang X, Shanmugam KT, Ingram LO (2011) L-malate production by metabolically engineered Escherichia coli. Appl Environ Microbiol 77:427–434
Zhang C, Zhou C, Assavasirijinda N, Yu B, Wang L, Ma Y (2017) Non-sterilized fermentation of high optically pure D-lactic acid by a genetically modified Thermophilic Bacillus coagulans strain. Microb Cell Factories 16:213
Zheng L, Xu T, Bai Z, He B (2014) Mn2+/Mg2+-dependent pyruvate kinase from a D-lactic acid-producing bacterium Sporolactobacillus inulinus: characterization of a novel Mn(2)(+)-mediated allosterically regulated enzyme. Appl Microbiol Biotechnol 98:1583–1593
Zhou S, Causey TB, Hasona A, Shanmugam KT, Ingram LO (2003) Production of optically pure D-lactic acid in mineral salts medium by metabolically engineered Escherichia coli W3110. Appl Environ Microbiol 69:399–407
Zhou L, Niu DD, Tian KM, Chen XZ, Prior BA, Shen W, Shi GY, Singh S, Wang ZX (2012) Genetically switched D-lactate production in Escherichia coli. Metab Eng 14:560–568
Zhu Y, Eiteman MA, DeWitt K, Altman E (2007) Homolactate fermentation by metabolically engineered Escherichia coli strains. Appl Environ Microbiol 73:456–464
Acknowledgments
This research was financially supported under One Research One Graduation (OROG) scholarship, Suranaree University of Technology, Thailand. Many thanks are also expressed to staff of the Office of International Relations at Ubon Ratchathani University and Dr. Bob Tremayne for their assistance with English.
Funding
This study was funded by Suranaree University of Technology, Thailand under One Research One Graduation (OROG-2559) scholarship.
Author information
Authors and Affiliations
Contributions
KJ conceived and designed research. SI, NW, and CP conducted experiments. PK contributed new reagents or analytical tools. KJ and SSJ analyzed data. SI and KJ wrote the manuscript. All authors read and approved the manuscript.
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Ethical approval
This article does not contain any studies with human participants or animals performed by any of the authors.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
In, S., Khunnonkwao, P., Wong, N. et al. Combining metabolic engineering and evolutionary adaptation in Klebsiella oxytoca KMS004 to significantly improve optically pure D-(−)-lactic acid yield and specific productivity in low nutrient medium. Appl Microbiol Biotechnol 104, 9565–9579 (2020). https://doi.org/10.1007/s00253-020-10933-0
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00253-020-10933-0