Abstract—
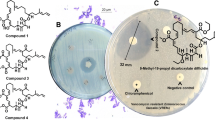
Planctomycetes are a phylogenetic group of bacteria with complex cell organization, large genomes and a wide spectrum of genome-encoded secondary metabolites. As suggested by analysis of available planctomycete genomes, these bacteria represent a promising source of potentially novel biologically active compounds. The number of cultured planctomycetes, however, remains limited. This study was undertaken in order to assess the presence of antimicrobial activity and its spectrum in the recently described freshwater planctomycete Lacipirellula parvula PX69T representing the novel family Lacipirellulaceae. The genome of strain PX69T contained 6 gene clusters encoding type III polyketide synthases, non-ribosomal peptide synthetases, and other secondary metabolites, which displayed low similarity to those in other bacteria. Strain PX69T was shown to exhibit antimicrobial activity against a number of test microorganisms. The highest antimicrobial activity, up to 83‒87% of growth inhibition, was observed against Staphylococcus aureus and Candida albicans. The medium composition and the cultivation time have been optimized in order to maximize antimicrobial activity of strain PX69T.





Similar content being viewed by others
REFERENCES
Boedeker, C., Schüler, M., Reintjes, G., Jeske, O., van Teeseling, M.C., Jogler, M., Rast, P., Borchert, D., Devos, D.P., Kucklick, M., Schaffer, M., Kolter, R., van Niftrik, L., Engelmann, S., Amann, R., et al., Determining the bacterial cell biology of Planctomycetes, Nat. Commun., 2017, vol. 8, pp. 14853. https://doi.org/10.1038/ncomms14853
Buckley, D.H., Huangyutitham, V., Nelson, T.A., Rumberger, A., and Thies, J.E., Diversity of Planctomycetes in soil in relation to soil history and environmental heterogeneity, Appl. Environ. Microbiol., 2006, vol. 72, pp. 4522–4531.
Cotter, P.D., Ross, R.P., and Hill, C., Bacteriocins ‒ a viable alternative to antibiotics?, Nat. Rev. Microbiol., 2013, vol. 11, pp. 95‒105.
Dedysh, S.N. and Ivanova, A.A., Planctomycetes in boreal and subarctic wetlands: diversity patterns and potential ecological functions, FEMS Microbiol. Ecol., 2019, vol. 95. fiy227. https://doi.org/10.1093/femsec/fiy227
Dedysh, S.N., Kulichevskaya, I.S., Beletsky, A.V., Ivanova, A.A., Rijpstra, W.I.C., Sinninghe Damsté, J.S., Mardanov, A.V., and Ravin, N.V., Lacipirellula parvula gen. nov., sp. nov., representing a lineage of planctomycetes widespread in low-oxygen habitats, description of the family Lacipirellulaceae fam. nov. and proposal of the orders Pirellulales ord. nov., Gemmatales ord. nov. and Isosphaerales ord. nov., Syst. Appl. Microbiol., 2020, vol. 43, art. 126050. https://doi.org/10.1016/j.syapm.2019.126050
Donadio, S., Monciardini, P., and Sosio, M., Polyketide synthases and nonribosomal peptide synthetases: the emerging view from bacterial genomics, Nat. Prod. Rep., 2007, vol. 24, pp. 1073‒1109. https://doi.org/10.1039/b514050c
Fischbach, M.A. and Walsh, C.T., Assembly-line enzymology for polyketide and nonribosomal peptide antibiotics: logic, machinery, and mechanisms, Chem. Rev., 2006, vol. 106, pp. 3468‒3496.
Fuerst, J.A., The planctomycetes: emerging models for microbial ecology, evolution and cell biology, Microbiology (SGM), 1995, vol. 141, pp. 1493–1506.
Glöckner, F.O., Kube, M., Bauer, M., Teeling, H., Lombardot, T., Ludwig, W., Gade, D., Beck, A., Borzym, K., Heitmann, K., Rabus, R., Schlesner, H., Amann, R., and Reinhardt, R., Complete genome sequence of the marine planctomycete Pirellula sp. strain 1, Proc. Natl. Acad. Sci. U. S. A., 2003, vol. 100, pp. 8298–8303.
Graça, A.P., Calisto, R., and Lage, O.M., Planctomycetes as novel source of bioactive molecules, Front. Microbiol. 2016, vol. 7, art. 1241. https://doi.org/10.3389/fmicb.2016.01241
Guo, M., Han, X., Jin, T., Zhou, L., Yang, J., Li, Z., Chen, J., Geng, B., Zou, Y., Wan, D., Li, D., Dai, W., Wang, H., Chen, Y., Ni, P., et al., Genome sequences of three species in the family Planctomycetaceae,J. Bacteriol., 2012, vol. 194, pp. 3740‒3741.
Jeske, O., Jogler, M., Petersen, J., Sikorski, J., and Jogler, C., From genome mining to phenotypic microarrays: Planctomycetes as source for novel bioactive molecules, Antonie van Leeuwenhoek, 2013, vol. 104, pp. 551‒567. https://doi.org/10.1007/s10482-013-0007-1
Jeske, O., Surup, F., Ketteniß, M., Rast, P., Förster, B., Jogler, M., Wink, J., and Jogler, C., Developing techniques for the utilization of Planctomycetes as producers of bioactive molecules, Front. Microbiol., 2016, vol. 7, art. 1242. https://doi.org/10.3389/fmicb.2016.01242
Ivanova, A.A., Naumoff, D.G., Miroshnikov, K.K., Liesack, W., and Dedysh, S.N., Comparative genomics of four Isosphaeraceae planctomycetes: a common pool of plasmids and glycoside hydrolase genes shared by Paludisphaera borealis PX4T, Isosphaera pallida IS1BT, Singulisphaera acidiphila DSM 18658T, and strain SH-PL62, Front. Microbiol., 2017, vol. 8, art. 412. https://doi.org/10.3389/fmicb.2017.00412
Lage, O.M. and Bondoso, J., Planctomycetes and macroalgae, a striking association, Front. Microbiol., 2014, vol. 5, art. 267. https://doi.org/10.3389/fmicb.2014.00267
Ravin, N.V., Rakitin, A.L., Ivanova, A.A., Beletsky, A.V., Kulichevskaya, I.S., Mardanov, A.V., and Dedysh, S.N., Genome analysis of Fimbriiglobus ruber SP5T, a planctomycete with confirmed chitinolytic capability, Appl. Environ. Microbiol., 2018, vol. 84, art. e02645-1. https://doi.org/10.1128/AEM.02645-17
Reintjes, G., Arnosti, C., Fuchs, B.M., and Amann, R., An alternative polysaccharide uptake mechanism of marine bacteria, ISME J., 2017, vol. 11, pp. 1640‒1650.
Schwarzer, D., Finking, R., and Marahiel, M.A., Nonribosomal peptides: from genes to products, Nat. Prod. Rep., 2003, vol. 20, pp. 275‒287.
Staley, J.T., Fuerst, J.A., Giovannoni, S., and Schlesner, H., The order Planctomycetales and the genera Planctomyces, Pirellula, Gemmata, and Isosphaera, in The Prokaryotes: A Handbook on the Biology of Bacteria: Ecophysiology, Isolation, Identification, Applications, Balows, A., Trüper, H.G., Dworkin, M., Harder, W., and Schleifer, K.-H., Eds., NY: Springer, 1992, pp. 3710–3731.
Tracanna, V., de Jong, A., Medema, M.H., and Kuipers, O.P., Mining prokaryotes for antimicrobial compounds: from diversity to function, FEMS Microbiol. Rev., 2017, vol. 41, pp. 417‒442.
Weber, T., Blin, K., Duddela, S., Krug, D., Kim, H.U., Bruccoleri, R., Lee, S.Y., Fischbach, M.A., Müler, R., Wohlleben, W., Breitling, R., Takano, E., and Medema, M.H., antiSMASH 3.0-a comprehensive resource for the genome mining of biosynthetic gene clusters, Nucl. Acids Res., 2015, vol. 43. W237–W243.
Wiegand, S., Jogler, M., and Jogler C., On the maverick Planctomycetes,FEMS Microbiol. Rev., 2018, vol. 42, pp. 739–760.
Wiegand, S., Jogler, M., Boedeker, C., Pinto, D., Vollmers, J., Rivas-Marín, E., Kohn, T., Peeters, S.H., Heuer, A., Rast, P., Oberbeckmann, S., Bunk, B., Jeske, O., Meyerdierks, A., Storesund, J.E., et al., Cultivation and functional characterization of 79 planctomycetes uncovers their unique biology, Nat. Microbiol., 2020, vol. 5, pp. 126‒140. https://doi.org/10.1038/s41564-019-0588-1
Yadav, S., Vaddavalli, R., Siripuram, S., Eedara, R.V.V., Yadav, S., Rabishankar, O., Lodha, T., Chintalapati, S., and Chintalapati, V., Planctopirus hydrillae sp. nov., an antibiotic producing planctomycete isolated from the aquatic plant Hydrilla and its whole genome shotgun sequence analysis, J. Antibiotics, 2018, vol. 71, pp. 575‒583.
Funding
The work was supported by the Russian Science Foundation, project no. 16-14-10210.
Author information
Authors and Affiliations
Contributions
The idea and scheme of the work were proposed by SD. Genome analysis for strain PX69Т was carried out by VS. Experiments on planctomycete cultivation, extraction of its secondary metabolites, and determination of their antimicrobial activity were carried out by SB and VS. SB and SD wrote the manuscript. All authors discussed the experimental results.
Corresponding author
Ethics declarations
The authors declare that they have no conflict of interest. This article does not contain any studies involving animals or human participants performed by any of the authors.
Additional information
Translated by P. Sigalevich
Rights and permissions
About this article
Cite this article
Belova, S.E., Saltykova, V.A. & Dedysh, S.N. Antimicrobial Activity of a Novel Freshwater Planctomycete Lacipirellula parvula PX69T . Microbiology 89, 503–509 (2020). https://doi.org/10.1134/S0026261720050045
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1134/S0026261720050045