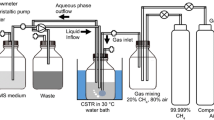
Abstract—
The composition of the microbial community formed in the course of long-term (over 60 days) continuous cultivation of an obligate methanotroph Methylococcus sp. Concept-8 on natural gas in a 50-L bioreactor at 42°C and pH 5.6 was monitored. Stable growth with high optical density of the culture (average OD540 = 13.5), specific growth rate of 0.2 h–1, and biomass yield of 4.0–4.5 g dry matter L–1 continued for 50 days. During the subsequent period of regime instability, optical density decreased sharply, down to OD540 = 4. Formate, acetate, and a number of unidentified fatty acids derived from oxidation of methane homologs were detected in the culture liquid. Microscopy revealed the highest share of satellite bacteria (10–15% of the total cell number) during the period of stable growth, while it dropped to 4.5% during growth decline. Molecular profiling of the community composition based on 16S rRNA gene sequence analysis revealed members of the genera Cohnella, Brevibacillus, Azospirillim, Thermomonas, Cupriavidus, and Paenibacillus among the major microbial satellites responsible for utilization of metabolites and the products of lysis of the methanotroph cells. During the phase of growth instability, the relative abundance of Brevibacillus decreased drastically; these organisms were most likely responsible for acetate oxidation. Isolates obtained by means of cultivation represented only a minor part of microbial diversity revealed in the community by molecular analysis. Isolation of the cultures of Brevibacillus and Cupriavidus species representing the functionally important components of the community opens the way for targeted construction of associations with strain Concept-8, which will display high productivity and stable growth.



Similar content being viewed by others
REFERENCES
Alves, M.P., Rainey, F.A., Nobre, M.F., and da Costa, M.S., Thermomonas hydrothermalis sp. nov., a new slightly thermophilic γ-proteobacterium isolated from a hot spring in Central Portugal, Syst. Appl. Microbiol., 2003, vol. 26, no. 1, pp. 70–75.
Bothe, H., Møller Jensen, K., Mergel, A., Larsen, J., Jørgensen, C., Bothe, H., and Jørgensen, L., Heterotrophic bacteria growing in association with Methylococcus capsulatus (Bath) in a single cell protein production process, Appl. Microbiol. Biotechnol., 2002, vol. 59, no. 1, pp. 33–39.
Butorova, I.A., Microbial community on natural gas and possibilities of its regulation, Extended abstract of Cand. Sci. (Biol.) Dissertation, Leningrad, 1991.
Galchenko, V.F., Metanotrofnye bakterii (Methanotrophic Bacteria), Moscow: GEOS, 2001.
Grigoryan, A.N. and Gorskaya, L. Ispolzovanie prirodnogo gaza dlya mikrobiologicheskogo sinteza (Use of Natural Gas for Microbiological Synthesis), Moscow: ONTI Microbioprom, 1970.
Hamer, G. and Harrison, D.E.F., Single cell protein: the technology, economics and future potential, in Hydrocarbons in Biotechnology, Harrison, D.E.F., Higgins, I.J., and London, W.R., Eds, London: Heyden Institute of Petroleum, 1980, pp. 59–73.
Hanson, R. and Hanson, T., Methanotrophic bacteria, Microbiol. Rev., 1996, vol. 60, no. 2, pp. 439–471.
Hartmann, A. and Zimmer, W., Physiology of Azospirillum, in Azospirillum/Plant Associations, Okon, Y., Ed., Boca Raton: CRC, 1994, pp. 15–39.
Ho, A., de Roy, K., Thas, O., de Neve, J., Hoefman, S., Vandamme, P., Heylen, K., and Boon, N., The more, the merrier: Heterotroph richness stimulates methanotrophic activity, ISME J., 2014, vol. 8, no. 9, pp. 1945–1948.
Iguchi, H., Yurimoto, H., and Sakai, Y., Stimulation of methanotrophic growth in cocultures by cobalamin excreted by rhizobia, Appl. Environ. Microbiol., 2011, vol. 77, no. 24, pp. 8509–8515.
Kalyuzhnaya, M.G., Yang, S., Rozova, O.N., Smalley, N.E., Clubb, J., Lamb, A., Gowda, G.A.N., Raftery, D., Fu, Y., Bringel, F., Vuilleumier, S., Beck, D.A.C., Trotsenko, Y.A., Khmelenina, V.N., and Lidstrom, M.E., Highly efficient methane biocatalysis revealed in a methanotrophic bacterium, Nat. Commun., 2013, vol. 4, pp. 1–7, art. 2785.
Kämpfer, P., Rosselló-Mora, R., Falsen, E., Busse, H.-J., and Tindall, B.J., Cohnella thermotolerans gen. nov., sp. nov., and classification of “Paenibacillus hongkongensis” as Cohnella hongkongensis sp. nov., Int. J. Syst. Evol. Microbiol., 2006, vol. 56, no. 4, pp. 781–786.
Khmelenina, V.N., Murrell, J.C., Smith, V.J., and Trotsenko, Y.A., Physiology and biochemistry of the aerobic methanotrophs, in Aerobic Utilization of Hydrocarbons, Oils and Lipids. Handbook of Hydrocarbon and Lipid Microbiology, Rojo, F., Ed., Cham: Springer, 2018, pp. 1–25.
Lalov, V.V., Analysis and synthesis of energotechnological systems for fodder protein production from natural gas, Extended abstract of Doctoral (Biol.) Dissertation, Moscow, 1991.
Leadbetter, E.R. and Forster, J.W., Studies on some methane-utilizing bacteria, Arch. Microbiol., 1958, vol. 30, pp. 91–118.
Linton, J.D. and Drozd, J.W., Microbial interactions and communities in biotechnology, in Microbial Interactions and Communities, Bull, A.T. and Slater, J.H., Eds., Academic Press: London, 1982, vol. 1, pp. 357–406.
Magoč, T. and Salzberg, S.L., FLASH: fast length adjustment of short reads to improve genome assemblies, Bioinformatics, 2011, vol. 27, no. 21, pp. 2957–2963.
Orlova, M.V., Tarlachkov, S.V., Dubinina, G.A., Belousova, E.V., Tutukina, M.N., and Grabovich, M.Y., Genomic insights into metabolic versatility of a lithotrophic sulfur-oxidizing diazotrophic Alphaproteobacterium Azospirillum thiophilum,FEMS Microbiol. Ecol., 2016, vol. 92, p. fiw199.
Oshkin, I.Yu., Khmelenina, V.N., But, S.Yu., Miroshnikov, K.K., Belova, S.E., Khokhlachev, N.S., Chernushkin, D.V., Beletsky, A.V., Mardanov, A.V., Ravin, N.V., Popov, V.O., Dedysh, S.N., and Pimenov, N.V., Analysis of the complete genome sequence of strain Concept-8, a novel representative of the genus Methylococcus,Microbiology (Moscow), 2020, vol. 89, no. 3, pp. 309–317.
Pruesse, E., Peplies, J., and Glöckner, F.O., SINA: accurate high-throughput multiple sequence alignment of ribosomal RNA genes, Bioinformatics, 2012, vol. 28, no. 14, pp. 1823–1829.
Smith, T.J. and Dalton, H., Biocatalysis by methane monooxygenase and its implications for the petroleum industry, in Petroleum Biotechnology: Developments and Perspective. Studies in Surface Science and Catalysis, Vazquez-Duhalt, R. and Quintero-Ramirez, R., Eds., Amsterdam: Elsevier, 2004, vol. 151, pp. 177–192.
Stock, M., Hoefman, S., Kerckhof, F.M., Boon, N., De Vos, P., De Baets, B., Heylen, K., and Waegeman, W., Exploration and prediction of interactions between methanotrophs and heterotrophs, Res. Microbiol., 2013, vol. 164, no. 10, pp. 1045–1054.
Strong, P.J., Xie, S., and Clarke, W.P., Methane as a resource: can the methanotrophs add value?, Environ. Sci. Technol., 2015, vol. 49, no. 7, pp. 4001–4018.
Tikhonova, E.N., Grouzdev, D.S., and Kravchenko, I.K., Azospirillum palustre sp. nov., a methylotrophic nitrogen-fixing species isolated from raised bog, Int. J. Syst. Evol. Microbiol., 2019, vol. 69, no. 9, pp. 2787–2793.
Trotsenko, Y.A. and Murrell, J.C., Metabolic aspects of aerobic obligate methanotrophy, Adv. Appl. Microbiol., 2008, vol. 63, pp. 183–229.
Weisburg, W.G., Barns, S.M., Pelletier, D.A., and Lane, D.J., 16S ribosomal DNA amplification for phylogenetic study, J. Bacteriol., 1991, vol. 173, pp. 697–703.
ACKNOWLEDGMENTS
The authors are grateful to V.V. Kevbrin for his assistance in identification of organic compounds in the culture liquid.
Funding
The work was supported by the Ministry of Education and Science of the Russian Federation, Federal Target Program grant no. 075-15-2019-1830, unique project ID RFMEFI60719X0297. Electron microscopy was performed at the Center for Collective Use of the Collection of Microorganisms UNIQEM, Federal Research Center for Biotechnology of the Russian Academy of Sciences. Molecular studies were carried out at the Collective Use Center “Bioengineering”, Federal Research Center for Biotechnology of the Russian Academy of Sciences.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interests. The authors declare that they have no conflict of interest.
The authors’ contribution. N.S. Khokhlachev, V.A. Se-menova, O.P. Chervyakova, and D.V. Chernushkin conducted continuous cultivation and growth monitoring of strain Concept-8. Sample collection and data analysis were performed by I.Yu. Oshkin. S.E. Belova was in charge of cell counts by microscopy and isolation of satellite bacteria. Electron microscopy was performed by E.N. Tikhonova. A.V. Mardanov and N.V. Ravin were in charge of DNA isolation, sequencing, and primary data analysis. The text of the manuscript was written by S.N. Dedysh, I.Yu. Oshkin, V.O. Popov, and N.V. Pimenov. All authors participated in the discussion of the results.
Statement on the welfare of animals. This article does not contain any studies involving animals or human participants performed by any of the authors.
Additional information
Translated by E. Makeeva
Rights and permissions
About this article
Cite this article
Oshkin, I.Y., Belova, S.E., Khokhlachev, N.S. et al. Molecular Analysis of the Microbial Community Developing in Continuous Culture of Methylococcus sp. Concept-8 on Natural Gas. Microbiology 89, 551–559 (2020). https://doi.org/10.1134/S0026261720050173
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1134/S0026261720050173