Unravelling Seascape Patterns of Cryptic Life Stages: Non-Reef Habitat Use in Juvenile Parrotfishes
Abstract
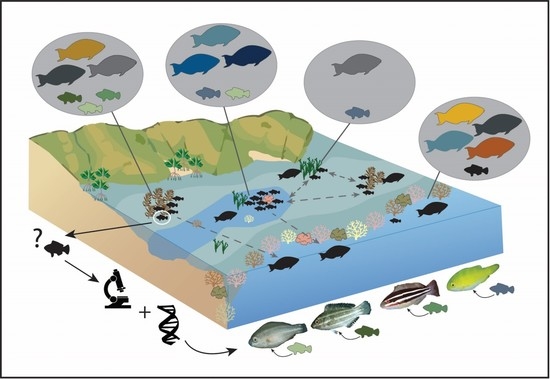
:1. Introduction
2. Materials and Methods
2.1. Study Site
2.2. Fish Surveys and Fish Collections
2.3. DNA Sequencing
2.4. Analysis of Sequences
3. Results
4. Discussion
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Nagelkerken, I.; Sheaves, M.; Baker, R.; Connolly, R.M. The seascape nursery: A novel spatial approach to identify and manage nurseries for coastal marine fauna. Fish Fish. 2015, 16, 362–371. [Google Scholar] [CrossRef] [Green Version]
- Beck, M.W.; HECK, K.L.; ABLE, K.W.; Childers, D.L.; Eggleston, D.B.; Gillanders, B.M.; Halpern, B.; Hays, C.G.; HOSHINO, K.; Minello, T.J.; et al. The Identification, Conservation, and Management of Estuarine and Marine Nurseries for Fish and Invertebrates. Bioscience 2001, 51, 633–641. [Google Scholar] [CrossRef]
- Lefcheck, J.S.; Hughes, B.B.; Johnson, A.J.; Pfirrman, B.W.; Rasher, D.B.; Smyth, A.R.; Williams, B.L.; Beck, M.W.; Orth, R.J. Are coastal habitats important nurseries? A meta-analysis. Conserv. Lett. 2019, e12645. [Google Scholar] [CrossRef]
- Sheaves, M.; Baker, R.; Nagelkerken, I.; Connolly, R.M. True value of estuarine and coastal nurseries for fish: Incorporating complexity and dynamics. Estuaries Coasts 2015, 38, 401–414. [Google Scholar] [CrossRef] [Green Version]
- Adams, A.J.; Dahlgren, C.P.; Kellison, G.T.; Kendall, M.S.; Layman, C.A.; Ley, J.A.; Nagelkerken, I.; Serafy, J.E. Nursery function of tropical back-reef systems. Mar. Ecol. Prog. Ser. 2006, 318, 287–301. [Google Scholar] [CrossRef]
- Dahlgren, C.P.; Kellison, T.G.; Adams, A.J.; Gillanders, B.M.; Kendall, M.S.; Layman, C.A.; Ley, J.A.; Nagelkerken, I.; Serafy, J.E. Marine nurseries and effective juvenile habitats: Concepts and applications. Mar. Ecol. Prog. Ser. 2006, 312, 291–295. [Google Scholar] [CrossRef] [Green Version]
- Nagelkerken, I.; van der Velde, G.; Gorissen, M.W.; Meijer, G.J.; van’t Hof, T.; den Hartog, C. Importance of mangroves, seagrass beds and the shallow coral reef as a nursery for important coral reef fishes, using a visual census technique. Estuar. Coast. Shelf Sci. 2000, 51, 31–44. [Google Scholar] [CrossRef]
- Fulton, C.J.; Berkström, C.; Wilson, S.K.; Abesamis, R.A.; Bradley, M.; Åkerlund, C.; Barrett, L.T.; Bucol, A.A.; Chacin, D.H.; Chong-seng, K.M.; et al. Macroalgal meadow habitats support fish and fisheries in diverse tropical seascapes. Fish Fish. 2020, 21, 1–18. [Google Scholar] [CrossRef]
- Dorenbosch, M.; Grol, M.G.G.; de Groene, A.; van Der Velde, G.; Nagelkerken, I. Piscivore assemblages and predation pressure affect relative safety of some back-reef habitats for juvenile fish in a Caribbean bay. Mar. Ecol. Prog. Ser. 2009, 379, 181–196. [Google Scholar] [CrossRef] [Green Version]
- Grol, M.G.G.; Nagelkerken, I.; Rypel, A.L.; Layman, C.A. Simple ecological trade-offs give rise to emergent cross-ecosystem distributions of a coral reef fish. Oecologia 2011, 165, 79–88. [Google Scholar] [CrossRef] [Green Version]
- Kimirei, I.A.; Nagelkerken, I.; Trommelen, M.; Blankers, P.; van Hoytema, N.; Hoeijmakers, D.; Huijbers, C.M.; Mgaya, Y.D.; Rypel, A.L. What drives ontogenetic niche shifts of fishes in coral reef ecosystems? Ecosystems 2013, 16, 783–796. [Google Scholar] [CrossRef] [Green Version]
- Dahlgren, C.P.; Eggleston, D.B. Ecological processes underlying ontogenetic habitat shifts in a coral reef fish. Ecology 2000, 81, 2227–2240. [Google Scholar] [CrossRef]
- Gillanders, B.M.; Able, K.W.; Brown, J.A.; Eggleston, D.B.; Sheridan, P.F. Evidence of connectivity between juvenile and adult habitats for mobile marine fauna: An important component of nurseries. Mar. Ecol. Prog. Ser. 2003, 247, 281–295. [Google Scholar] [CrossRef] [Green Version]
- Galaiduk, R.; Radford, B.T.; Saunders, B.J.; Newman, S.J.; Harvey, E.S. Characterizing ontogenetic habitat shifts in marine fishes: Advancing nascent methods for marine spatial management. Ecol. Appl. 2017, 27, 1776–1788. [Google Scholar] [CrossRef]
- Bellwood, D.R. Ontogenetic changes in the diet of early post-settlement Scarus species (Pisces: Scaridae). J. Fish Biol. 1988, 33, 213–219. [Google Scholar] [CrossRef]
- Clements, K.D.; German, D.P.; Piché, J.; Tribollet, A.; Choat, J.H. Integrating ecological roles and trophic diversification on coral reefs: Multiple lines of evidence identify parrotfishes as microphages. Biol. J. Linn. Soc. 2017, 120, 729–751. [Google Scholar] [CrossRef]
- Chen, L.-S. Post-settlement diet shift of Chlorurus sordidus and Scarus schlegeli (Pisces: Scaridae). Zool. Stud. 2002, 41, 47–58. [Google Scholar]
- Streit, R.P.; Bellwood, D.R. High prevalence of homing behaviour among juvenile coral-reef fishes and the role of body size. Coral Reefs 2017, 36, 1083–1095. [Google Scholar] [CrossRef]
- Welsh, J.Q.; Goatley, C.H.R.R.; Bellwood, D.R. The ontogeny of home ranges: Evidence from coral reef fishes. Proc. R. Soc. B Biol. Sci. 2013, 280, 1–7. [Google Scholar] [CrossRef] [Green Version]
- Huijbers, C.M.; Nagelkerken, I.; Layman, C.A. Fish movement from nursery bays to coral reefs: A matter of size? Hydrobiologia 2015, 750, 89–101. [Google Scholar] [CrossRef] [Green Version]
- Kimirei, I.A.; Nagelkerken, I.; Griffioen, B.; Wagner, C.; Mgaya, Y.D. Ontogenetic habitat use by mangrove/seagrass-associated coral reef fishes shows flexibility in time and space. Estuar. Coast. Shelf Sci. 2011, 92, 47–58. [Google Scholar] [CrossRef]
- Van Lier, J.R.; Wilson, S.K.; Depczynski, M.; Wenger, L.N.; Fulton, C.J. Habitat connectivity and complexity underpin fish community structure across a seascape of tropical macroalgae meadows. Landsc. Ecol. 2018, 33, 1287–1300. [Google Scholar] [CrossRef]
- Nagelkerken, I.; Bothwell, J.; Nemeth, R.S.; Pitt, J.M.; van der Velde, G. Interlinkage between Caribbean coral reefs and seagrass beds through feeding migrations by grunts (Haemulidae) depends on habitat accessibility. Mar. Ecol. Prog. Ser. 2008, 368, 155–164. [Google Scholar] [CrossRef] [Green Version]
- Berkström, C.; Eggertsen, L.; Goodell, W.; Cordeiro, C.A.M.M.M.M.; Lucena, M.B.; Gustafsson, R.; Bandeira, S.; Jiddawi, N.; Ferreira, C.E.L.E.L. Thresholds in seascape connectivity: The spatial arrangement of nursery habitats structure fish communities on nearby reefs. Ecography 2020, 43, 882–896. [Google Scholar] [CrossRef]
- Turgeon, K.; Robillard, A.; Grégoire, J.; Duclos, V.; Kramer, D.L. Functional connectivity from a reef fish perspective: Behavioral tactics for moving in a fragmented landscape. Ecology 2010, 91, 3332–3342. [Google Scholar] [CrossRef]
- Hitt, S.; Pittman, S.J.; Nemeth, R.S. Diel movements of fishes linked to benthic seascape structure in a Caribbean coral reef ecosystem. Mar. Ecol. Prog. Ser. 2011, 427, 275–291. [Google Scholar] [CrossRef] [Green Version]
- Nagelkerken, I.; Roberts, C.M.; van der Velde, G.; Dorenbosch, M.; van Riel, M.C.; Cocheret de la Morinière, E.; Nienhuis, P.H. How important are mangroves and seagrass beds for coral-reef fish? The nursery hypothesis tested on an island scale. Mar. Ecol. Prog. Ser. 2002, 244, 299–305. [Google Scholar] [CrossRef] [Green Version]
- Berkström, C.; Gullström, M.; Lindborg, R.; Mwandya, A.W.; Yahya, S.A.S.; Kautsky, N.; Nyström, M. Exploring ‘knowns’ and ‘unknowns’ in tropical seascape connectivity with insights from East African coral reefs. Estuar. Coast. Shelf Sci. 2012, 107, 1–21. [Google Scholar] [CrossRef]
- Lugendo, B.R.; Nagelkerken, I.; van der Velde, G.; Mgaya, Y.D. The importance of mangroves, mud and sand flats, and seagrass beds as feeding areas for juvenile fishes in Chwaka Bay, Zanzibar: Gut content and stable isotope analyses. J. Fish Biol. 2006, 69, 1639–1661. [Google Scholar] [CrossRef]
- Sambrook, K.; Hoey, A.S.; Andréfouët, S.; Cumming, G.S.; Duce, S.; Bonin, M.C. Beyond the reef: The widespread use of non-reef habitats by coral reef fishes. Fish Fish. 2019, 20, 1–18. [Google Scholar] [CrossRef]
- Honda, K.; Uy, W.H.; Baslot, D.I.; Pantallano, A.D.S.; Nakamura, Y.; Nakaoka, M. Diel habitat-use patterns of commercially important fishes in a marine protected area in the Philippines. Aquat. Biol. 2016, 24, 163–174. [Google Scholar] [CrossRef] [Green Version]
- Gullström, M.; Berkström, C.; Öhman, M.C.; Bodin, M.; Dahlberg, M. Scale-dependent patterns of variability of a grazing parrotfish (Leptoscarus vaigiensis) in a tropical seagrass-dominated seascape. Mar. Biol. 2011, 158, 1483–1495. [Google Scholar] [CrossRef]
- Nakamura, Y.; Horinouchi, M.; Shibuno, T.; Tanaka, Y.; Miyajima, T.; Koike, I.; Kurokura, H.; Sano, M. Evidence of ontogenetic migration from mangroves to coral reefs by black-tail snapper Lutjanus fulvus: Stable isotope approach. Mar. Ecol. Prog. Ser. 2008, 355, 257–266. [Google Scholar] [CrossRef]
- Bonaldo, R.M.; Hoey, A.S.; Bellwood, D.R. The Ecosystem Roles of Parrotfishes on Tropical Reefs. Oceanogr. Mar. Biol. Annu. Rev. 2014, 52, 81–132. [Google Scholar] [CrossRef]
- Bellwood, D.R.; Hoey, A.S.; Hughes, T.P. Human activity selectively impacts the ecosystem roles of parrotfishes on coral reefs. Proc. R. Soc. B Biol. Sci. 2012, 279, 1621–1629. [Google Scholar] [CrossRef] [Green Version]
- Taylor, B.M.; Houk, P.; Russ, G.R.; Choat, J.H. Life histories predict vulnerability to overexploitation in parrotfishes. Coral Reefs 2014, 33, 869–878. [Google Scholar] [CrossRef]
- Fulton, C.J.; Abesamis, R.A.; Berkström, C.; Depczynski, M.; Graham, N.A.J.; Holmes, T.H.; Kulbicki, M.; Noble, M.M.; Radford, B.T.; Tano, S.; et al. Form and Function of tropical macroalgal reefs in the Anthropocene. Funct. Ecol. 2019, 33, 989–999. [Google Scholar] [CrossRef]
- Igulu, M.M.; Nagelkerken, I.; Dorenbosch, M.; Grol, M.G.G.; Harborne, A.R.; Kimirei, I.A.; Mumby, P.J.; Olds, A.D.; Mgaya, Y.D. Mangrove habitat use by juvenile reef fish: Meta-analysis reveals that tidal regime matters more than biogeographic region. PLoS ONE 2014, 9, e114715. [Google Scholar] [CrossRef] [Green Version]
- Dorenbosch, M.; Grol, M.G.G.; Christianen, M.J.A.; Nagelkerken, I.; van Der Velde, G. Indo-Pacific seagrass beds and mangroves contribute to fish density and diversity on adjacent coral reefs. Mar. Ecol. Prog. Ser. 2005, 302, 63–76. [Google Scholar] [CrossRef] [Green Version]
- Bellwood, D.R.; Choat, J.H. A description of the juvenile phase colour pattern of 24 parrotfish species (family Scaridae) from the Great Barrier Reef, Australia. Rec. Aust. Mus. 1989, 41, 1–41. [Google Scholar] [CrossRef] [Green Version]
- Grober-Dunsmore, R.; Frazer, T.K.; Lindberg, W.J.; Beets, J. Reef fish and habitat relationships in a Caribbean seascape: The importance of reef context. Coral Reefs 2007, 26, 201–216. [Google Scholar] [CrossRef]
- Tano, S.; Eggertsen, M.; Wikström, S.A.; Berkström, C.; Buriyo, A.S.; Halling, C. Tropical seaweed beds are important habitats for mobile invertebrate epifauna. Estuar. Coast. Shelf Sci. 2016, 183, 1–12. [Google Scholar] [CrossRef]
- Eggertsen, L.; Ferreira, C.E.L.; Fontoura, L.; Kautsky, N.; Gullström, M.; Berkström, C. Seaweed beds support more juvenile reef fish than seagrass beds in a south-western Atlantic tropical seascape. Estuar. Coast. Shelf Sci. 2017, 196, 97–108. [Google Scholar] [CrossRef] [Green Version]
- Bradley, M.; Baker, R.; Nagelkerken, I.; Sheaves, M. Context is more important than habitat type in determining use by juvenile fish. Landsc. Ecol. 2019, 34, 427–442. [Google Scholar] [CrossRef]
- Feitosa, J.L.L.; Ferreira, B.P. Distribution and feeding patterns of juvenile parrotfish on algal-dominated coral reefs. Mar. Ecol. 2014, 36, 462–474. [Google Scholar] [CrossRef]
- Weigt, L.A.; Baldwin, C.C.; Driskell, A.; Smith, D.G.; Ormos, A.; Reyier, E.A. Using DNA barcoding to assess Caribbean reef rish biodiversity: Expanding taxonomic and geographic coverage. PLoS ONE 2012, 7, e41059. [Google Scholar] [CrossRef] [Green Version]
- De Leon, R.O.D.; White, A.T. Mangrove Rehabilitation in the Philippines. In An International Perspective on Wetland Rehabilitation; Streever, W., Ed.; Springer: Dordrecht, The Netherlands, 1999; pp. 37–42. ISBN 978-94-011-4683-8. [Google Scholar]
- Ward, R.D.; Zemlak, T.S.; Innes, B.H.; Last, P.R.; Hebert, P.D.N. DNA barcoding Australia’s fish species. Philisophical Trans. R. Soc. 2005, 1847–1857. [Google Scholar] [CrossRef]
- Costa, F.O.; Landi, M.; Martins, R.; Costa, M.H.; Costa, M.E.; Carneiro, M.; Alves, M.J.; Steinke, D.; Carvalho, G.R. A ranking system for reference libraries of DNA barcodes: Application to marine fish species from Portugal. PLoS ONE 2012, 7, e0035858. [Google Scholar] [CrossRef] [Green Version]
- McClure, E.C.; Sievers, K.T.; Abesamis, R.A.; Hoey, A.S.; Alcala, A.C.; Russ, G.R. Higher fish biomass inside than outside marine protected areas despite typhoon impacts in a complex reefscape. Biol. Conserv. 2020, 241, 108354. [Google Scholar] [CrossRef]
- Russ, G.R.; Questel, S.L.A.; Rizzari, J.R.; Alcala, A.C. The parrotfish–coral relationship: Refuting the ubiquity of a prevailing paradigm. Mar. Biol. 2015, 162, 2029–2045. [Google Scholar] [CrossRef]
- Stockwell, B.; Jadloc, C.R.L.; Abesamis, R.A.; Alcala, A.C.; Russ, G.R. Trophic and benthic responses to no-take marine reserve protection in the Philippines. Mar. Ecol. Prog. Ser. 2009, 389, 1–15. [Google Scholar] [CrossRef] [Green Version]
- Siqueira, A.C.; Bellwood, D.R.; Cowman, P.F. The evolution of traits and functions in herbivorous coral reef fishes through space and time. Proc. R. Soc. B Biol. Sci. 2019, 286, 20182672. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Russ, G.R. Distribution and abundance of herbivorous grazing fishes in the central Great Barrier Reef. I. Levels of variability across the entire continental shelf. Mar. Ecol. Prog. Ser. 1984, 20, 23–34. [Google Scholar] [CrossRef]
- Russ, G.R. Distribution and abundance of herbivorous grazing fishes in the central Great Barrier Reef. II. Patterns of zonation of mid-shelf and outhershelf reefs. Mar. Ecol. Prog. Ser. 1984, 20, 35–44. [Google Scholar] [CrossRef]
- Bellwood, D.R.; Tebbett, S.B.; Bellwood, O.; Mihalitsis, M.; Morais, R.A.; Streit, R.P.; Fulton, C.J. The role of the reef flat in coral reef trophodynamics: Past, present, and future. Ecol. Evol. 2018, 8, 4108–4119. [Google Scholar] [CrossRef]
- Dulvy, N.K.; Sadovy, Y.; Reynolds, J.D. Extinction vulnerability in marine populations. Fish Fish. 2003, 4, 25–64. [Google Scholar] [CrossRef]
- Lavergne, S.; Thuiller, W.; Molina, J.; Debussche, M. Environmental and human factors influencing rare plant local occurrence, extinction and persistence: A 115-year study in the Mediterranean region. J. Biogeogr. 2005, 32, 799–811. [Google Scholar] [CrossRef]
- Hubert, N.; Meyer, C.P.; Bruggemann, H.J.; Guérin, F.; Komeno, R.J.L.; Espiau, B.; Causse, R.; Williams, J.T.; Planes, S. Cryptic diversity in Indo-Pacific coral-reef fishes revealed by DNA-barcoding provides new support to the centre-of-overlap hypothesis. PLoS ONE 2012, 7, e28987. [Google Scholar] [CrossRef]
- Hubert, N.; Hanner, R. DNA Barcoding, species delineation and taxonomy: A historical perspective. DNA Barcodes 2015, 3, 44–58. [Google Scholar] [CrossRef]
- Cocheret de la Morinière, E.; Pollux, B.J.A.; Nagelkerken, I.; Hemminga, M.A.; Huiskes, A.H.L.; van der Velde, G. Ontogenetic dietary changes of coral reef fishes in the mangrove-seagrass-reef continuum: Stable isotopes and gut-content analysis. Mar. Ecol. Prog. Ser. 2003, 246, 279–289. [Google Scholar] [CrossRef] [Green Version]
- Mumby, P.J. Connectivity of reef fish between mangroves and coral reefs: Algorithms for the design of marine reserves at seascape scales at seascape scales. Biol. Conserv. 2006, 215–222. [Google Scholar] [CrossRef]
- Olds, A.D.; Albert, S.; Maxwell, P.S.; Pitt, K.A.; Connolly, R.M. Mangrove-reef connectivity promotes the effectiveness of marine reserves across the western Pacific. Glob. Ecol. Biogeogr. 2013, 22, 1040–1049. [Google Scholar] [CrossRef] [Green Version]
- Martin, T.S.H.; Olds, A.D.; Pitt, K.A.; Johnston, A.B.; Butler, I.R.; Maxwell, P.S.; Connolly, R.M. Effective protection of fish on inshore coral reefs depends on the scale of mangrove-reef connectivity. Mar. Ecol. Prog. Ser. 2015, 527, 157–165. [Google Scholar] [CrossRef] [Green Version]
- Olds, A.D.; Connolly, R.M.; Pitt, K.A.; Maxwell, P.S. Primacy of seascape connectivity effects in structuring coral reef fish assemblages. Mar. Ecol. Prog. Ser. 2012, 462, 191–203. [Google Scholar] [CrossRef] [Green Version]
- Olds, A.D.; Connolly, R.M.; Pitt, K.A.; Maxwell, P.S. Habitat connectivity improves reserve performance. Conserv. Lett. 2012, 5, 56–63. [Google Scholar] [CrossRef]
- Lugendo, B.R.; Nagelkerken, I.; Kruitwagen, G.; van der Velde, G.; Mgaya, Y.D. Relative importance of mangroves as feeding habitat for juvenile fish: A comparative study on mangrove habitats with different settings. Bull. Mar. Sci. 2007, 80, 497–512. [Google Scholar]
- Adams, A.J.; Ebersole, J.P. Use of back-reef and lagoon habitats by coral reef fishes. Mar. Ecol. Prog. Ser. 2002, 228, 213–226. [Google Scholar] [CrossRef] [Green Version]
- Mellin, C.; Kulbicki, M.; Ponton, D. Seasonal and ontogenetic patterns of habitat use in coral reef fish juveniles. Estuar. Coast. Shelf Sci. 2007, 75, 481–491. [Google Scholar] [CrossRef]
- Cocheret De La Morinière, E.; Pollux, B.J.A.; Nagelkerken, I.; van der Velde, G. Post-settlement life cycle migration patterns and habitat preference of coral reef fish that use seagrass and mangrove habitats as nurseries. Estuar. Coast. Shelf Sci. 2002, 55, 309–321. [Google Scholar] [CrossRef] [Green Version]
- Nagelkerken, I.; Dorenbosch, M.; Verberk, W.C.E.P.; De Morinière, E.C.; van der Velde, G.; Cocheret de la Morinière, E.; van der Velde, G. Importance of shallow-water biotopes of a Caribbean bay for juvenile coral reef fishes: Patterns in biotope association, community structure and spatial distribution. Mar. Ecol. Prog. Ser. 2000, 202, 175–192. [Google Scholar] [CrossRef] [Green Version]
- Fong, C.R.; Chancellor, K.S.; Renzi, J.J.; Robinson, D.R.; Barber, P.H.; Habtes, S.Y.; Fong, P. Epibionts on Turbinaria ornata, a secondary foundational macroalga on coral reefs, provide diverse trophic support to fishes. Mar. Environ. Res. 2018, 141, 39–43. [Google Scholar] [CrossRef]
- Johnson, G.B.; Taylor, B.M.; Robbins, W.D.; Franklin, E.C.; Toonen, R.; Bowen, B.; Choat, J.H. Diversity and Structure of Parrotfish Assemblages across the Northern Great Barrier Reef. Diversity 2019, 11, 14. [Google Scholar] [CrossRef] [Green Version]
- Welsh, J.Q.; Bellwood, D.R. How far do schools of roving herbivores rove? A case study using Scarus rivulatus. Coral Reefs 2012, 31, 991–1003. [Google Scholar] [CrossRef]
- Tolimieri, N. The relationship among microhabitat characteristics, recruitment and adult abundance in the stoplight parrotfish, Sparisoma Viride, at three spatial scales. Bull. Mar. Sci. 1998, 62, 253–268. [Google Scholar]
- Sale, P.F.; Danilowicz, B.S.; Doherty, P.J.; Williams, D.M.B. The relation of microhabitat to variation in recruitment of young-of-year coral reef fishes. Bull. Mar. Sci. 2005, 76, 123–142. [Google Scholar]
- Williams, D.M. Dynamics of the Pomacentrid community on small patch reefs in One Tree lagoon (Great Barrier Reef). Bull. Mar. Sci. 1980, 30, 159–170. [Google Scholar]
- Taylor, B.M.; Lindfield, S.J.; Choat, J.H. Hierarchical and scale-dependent effects of fishing pressure and environment on the structure and size distribution of parrotfish communities. Ecography 2015, 38, 520–530. [Google Scholar] [CrossRef]
- Abesamis, R.A.; Saenz-Agudelo, P.; Berumen, M.L.; Bode, M.; Jadloc, C.R.L.; Solera, L.A.; Villanoy, C.L.; Bernardo, L.P.C.; Alcala, A.C.; Russ, G.R. Reef-fish larval dispersal patterns validate no-take marine reserve network connectivity that links human communities. Coral Reefs 2017, 1–11. [Google Scholar] [CrossRef]
- Taylor, B.M.; Choat, J.H. Comparative demography of commercially important parrotfish species from Micronesia. J. Fish Biol. 2014, 84, 383–402. [Google Scholar] [CrossRef]
- Lou, D.C. Age Specific Patterns of Growth and Reproduction in Tropical Herbivorous Fishes. Ph.D. Thesis, James Cook University, Douglas, Australia, 1992. [Google Scholar]
- Nicholson, G.M.; Clements, K.D. Resolving resource partitioning in parrotfishes (Scarini) using microhistology of feeding substrata. Coral Reefs 2020, 39. [Google Scholar] [CrossRef]
- Bonaldo, R.M.; Bellwood, D.R. Size-dependent variation in the functional role of the parrotfish Scarus rivulatus on the Great Barrier Reef, Australia. Mar. Ecol. Prog. Ser. 2008, 360, 237–244. [Google Scholar] [CrossRef]
- Davis, K.; Carlson, P.M.; Lowe, C.G.; Warner, R.R.; Caselle, J.E. Parrotfish movement patterns vary with spatiotemporal scale. Mar. Ecol. Prog. Ser. 2017, 577, 149–164. [Google Scholar] [CrossRef] [Green Version]
- Huijbers, C.M.; Nagelkerken, I.; Debrot, A.O.; Jongejans, E. Geographic coupling of juvenile and adult habitat shapes spatial population dynamics of a coral reef fish. Ecology 2013, 94, 1859–1870. [Google Scholar] [CrossRef] [Green Version]
- Moffitt, E.A.; Botsford, L.W.; Kaplan, D.M.; O’Farrell, M.R. Marine reserve networks for species that move within a home range. Ecol. Appl. 2009, 19, 1835–1847. [Google Scholar] [CrossRef]
- Grüss, A.; Kaplan, D.M.; Guénette, S.; Roberts, C.M.; Botsford, L.W. Consequences of adult and juvenile movement for marine protected areas. Biol. Conserv. 2011, 144, 692–702. [Google Scholar] [CrossRef]
- Weeks, R. Incorporating seascape connectivity in conservation prioritisation. PLoS ONE 2017, 12, e0182396. [Google Scholar] [CrossRef] [PubMed]





| High | Med | Low | ||
|---|---|---|---|---|
| DNA Sequence Quality | ||||
| High Quality Bases | >95% | 80–95% | <80% | |
| Alignment | >90% | 50–90% | <50% | |
| Ambiguities | <5 | 5–15 | >15 | |
| Sequence Match on Databank | ||||
| % Similarity | >98% | 90–98% | <90% | |
| Databank References | ||||
| Peer review and accessibility | Published, accessible | Published, not accessible | Not published, not accessible | |
| Photo | Yes | No | Wrong photo | |
| Collection Details (e.g., lat/long, collector, and identifier) | Full details | Minimal details | No details | |
| Consistency of top hits | Top 10 all same species | 5 of 10 top hits same species | Mix of species for top 10 hits | |
| Adult Presence | Juvenile Presence | Juvenile Count | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Species | All | CR | MA | SG | LAG | All | MA | SG | LAG | MA | SG | LAG | |
| 1 | Calotomus carolinus | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| 2 | Calotomus spinidens | 1 | 0 | 1 | 1 | 0 | 1 | 1 | 0 | 0 | 4 | 0 | 0 |
| 3 | Cetoscarus ocellatus | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| 4 | Chlorurus bleekeri | 1 | 1 | 1 | 0 | 1 | 1 | 1 | 0 | 1 | 2 | 0 | 1 |
| 5 | Chlorurus bowersi/bleekeri | 0 | 0 | 0 | 0 | 0 | 1 | 1 | 0 | 0 | 6 | 0 | 0 |
| 6 | Chlorurus japanensis/capistratoides | 0 | 0 | 0 | 0 | 0 | 1 | 1 | 0 | 0 | 1 | 0 | 0 |
| 7 | Chlorurus spilurus | 1 | 1 | 1 | 0 | 1 | 1 | 1 | 0 | 0 | 4 | 0 | 0 |
| 8 | Hipposcarus longiceps | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| 9 | Leptoscarus vaigiensis | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| 10 | Scarus altipinnis | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| 11 | Scarus chameleon | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 0 | 2 | 0 | 0 |
| 12 | Scarus dimidiatus | 1 | 1 | 0 | 0 | 1 | 1 | 1 | 1 | 1 | 2 | 5 | 20 |
| 13 | Scarus flavipectoralis | 1 | 1 | 1 | 0 | 1 | 1 | 1 | 0 | 0 | 1 | 0 | 0 |
| 14 | Scarus forsteni | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| 15 | Scarus ghobban | 1 | 1 | 0 | 0 | 1 | 1 | 1 | 1 | 0 | 1 | 2 | 0 |
| 16 | Scarus hypselopterus | 1 | 1 | 1 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| 17 | Scarus niger | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| 18 | Scarus psittacus | 0 | 0 | 0 | 0 | 0 | 1 | 1 | 1 | 1 | 105 | 1 | 1 |
| 19 | Scarus quoyi | 1 | 1 | 0 | 0 | 0 | 1 | 1 | 1 | 1 | 19 | 2 | 2 |
| 20 | Scarus rivulatus | 1 | 1 | 0 | 0 | 0 | 1 | 1 | 0 | 1 | 41 | 0 | 3 |
| 21 | Scarus rubroviolaceus | 0 | 0 | 0 | 0 | 0 | 1 | 1 | 0 | 0 | 7 | 0 | 0 |
| 22 | Scarus schlegeli | 1 | 1 | 1 | 0 | 0 | 1 | 1 | 0 | 0 | 1 | 0 | 0 |
| 23 | Scarus spinus | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 1 | 0 | 0 | 0 |
| 24 | Scarus tricolor | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 1 |
| Total | 19 | 16 | 8 | 4 | 7 | 15 | 14 | 4 | 6 | 197 | 11 | 28 | |
| Count | Species | High | Med | Low | Total | TL Range (mm) |
|---|---|---|---|---|---|---|
| 1 | Scarus psittacus | 80 | 24 | 3 | 107 | 18–51 |
| 2 | Scarus rivulatus | 1 | 35 | 8 | 44 | 13–57 |
| 3 | Scarus dimidiatus | 27 | 27 | 32–98 | ||
| 4 | Scarus quoyi | 15 | 6 | 2 | 23 | 17–87 |
| 5 | Scarus rubroviolaceus | 7 | 7 | 24–75 | ||
| 6 | Chlorurus bowersi/bleekeri | 4 | 2 | 6 | 21–39 | |
| 7 | Calotomus spinidens | 4 | 4 | 36–42 | ||
| 8 | Chlorurus spilurus | 4 | 4 | 22–67 | ||
| 9 | Chlorurus bleekeri | 1 | 2 | 3 | 29–38 | |
| 10 | Scarus ghobban | 2 | 1 | 3 | 46–118 | |
| 11 | Scarus chameleon | 2 | 2 | 29–38 | ||
| 12 | Scarus flavipectoralis | 2 | 2 | 31 | ||
| 13 | Chlorurus japanensis/capistratoides | 1 | 1 | 29 | ||
| 14 | Scarus schlegeli | 1 | 1 | 39 | ||
| 15 | Scarus spinus | 1 | 1 | 21 | ||
| Grand Total | 112 | 82 | 15 | 209 |
| Juvenile | Adult | |||||||
|---|---|---|---|---|---|---|---|---|
| Species | CR | MA | LAG | SG | CR | MA | LAG | SG |
| Cetoscarus ocellatus | 1 | 1 | ||||||
| Chlorurus microrhinos | 1 | |||||||
| Scarus dimidiatus | 1 | 1 | 1 | 1 | ||||
| Scarus niger | 1 | 1 | 1 | |||||
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Sievers, K.T.; Abesamis, R.A.; Bucol, A.A.; Russ, G.R. Unravelling Seascape Patterns of Cryptic Life Stages: Non-Reef Habitat Use in Juvenile Parrotfishes. Diversity 2020, 12, 376. https://doi.org/10.3390/d12100376
Sievers KT, Abesamis RA, Bucol AA, Russ GR. Unravelling Seascape Patterns of Cryptic Life Stages: Non-Reef Habitat Use in Juvenile Parrotfishes. Diversity. 2020; 12(10):376. https://doi.org/10.3390/d12100376
Chicago/Turabian StyleSievers, Katie T., Rene A. Abesamis, Abner A. Bucol, and Garry R. Russ. 2020. "Unravelling Seascape Patterns of Cryptic Life Stages: Non-Reef Habitat Use in Juvenile Parrotfishes" Diversity 12, no. 10: 376. https://doi.org/10.3390/d12100376