Abstract
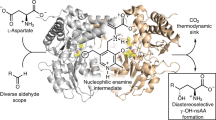
An ongoing challenge in chemical research is to design catalysts that select the outcomes of the reactions of complex molecules. Chemists rely on organocatalysts or transition metal catalysts to control stereoselectivity, regioselectivity and periselectivity (selectivity among possible pericyclic reactions). Nature achieves these types of selectivity with a variety of enzymes such as the recently discovered pericyclases—a family of enzymes that catalyse pericyclic reactions1. Most characterized enzymatic pericyclic reactions have been cycloadditions, and it has been difficult to rationalize how the observed selectivities are achieved2,3,4,5,6,7,8,9,10,11,12,13. Here we report the discovery of two homologous groups of pericyclases that catalyse distinct reactions: one group catalyses an Alder-ene reaction that was, to our knowledge, previously unknown in biology; the second catalyses a stereoselective hetero-Diels–Alder reaction. Guided by computational studies, we have rationalized the observed differences in reactivities and designed mutant enzymes that reverse periselectivities from Alder-ene to hetero-Diels–Alder and vice versa. A combination of in vitro biochemical characterizations, computational studies, enzyme co-crystal structures, and mutational studies illustrate how high regioselectivity and periselectivity are achieved in nearly identical active sites.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout



Similar content being viewed by others
Data availability
Data that support the findings of this study are available within the paper and its Supplementary Information, or are available from the corresponding author upon request. The atomic coordinates of PdxI, PdxI with 5, PdxI with 8, HpiI and HpiI with 5 have been deposited in the Protein Data Bank (http://www.rcsb.org) under accession codes 7BQJ, 7BQK, 7BQL, 7BQP and 7BQO, respectively.
References
Jamieson, C. S., Ohashi, M., Liu, F., Tang, Y. & Houk, K. N. The expanding world of biosynthetic pericyclases: Cooperation of experiment and theory for discovery. Nat. Prod. Rep. 36, 698–713 (2019).
Kim, H. J., Ruszczycky, M. W., Choi, S., Liu, Y. & Liu, H. Enzyme-catalysed [4+2] cycloaddition is a key step in the biosynthesis of spinosyn A. Nature 473, 109–112 (2011).
Wever, W. J. et al. Chemoenzymatic synthesis of thiazolyl peptide natural products featuring an enzyme-catalyzed formal [4 + 2] cycloaddition. J. Am. Chem. Soc. 137, 3494–3497 (2015).
Ohashi, M. et al. SAM-dependent enzyme-catalysed pericyclic reactions in natural product biosynthesis. Nature 549, 502–506 (2017).
Bailey, S. S. et al. Enzymatic control of cycloadduct conformation ensures reversible 1,3-dipolar cycloaddition in a prFMN-dependent decarboxylase. Nat. Chem. 11, 1049–1057 (2019).
Chen, Q. et al. Enzymatic intermolecular hetero-Diels-Alder reaction in the biosynthesis of tropolonic sesquiterpenes. J. Am. Chem. Soc. 141, 14052–14056 (2019).
Zhang, B. et al. Enzyme-catalysed [6+4] cycloadditions in the biosynthesis of natural products. Nature 568, 122–126 (2019).
Little, R. et al. Unexpected enzyme-catalysed [4+2] cycloaddition and rearrangement in polyether antibiotic biosynthesis. Nat. Catal. 2, 1045–1054 (2019).
Dan, Q. et al. Fungal indole alkaloid biogenesis through evolution of a bifunctional reductase/Diels–Alderase. Nat. Chem. 11, 972–980 (2019).
Zhang, Z. et al. Enzyme-catalyzed inverse-electron demand Diels–Alder reaction in the biosynthesis of antifungal ilicicolin H. J. Am. Chem. Soc. 141, 5659–5663 (2019).
Gustin, D. J. et al. Heavy atom isotope effects reveal a highly polarized transition state for chorismate mutase. J. Am. Chem. Soc. 121, 1756–1757 (1999).
DeClue, M. S., Baldridge, K. K., Künzler, D. E., Kast, P. & Hilvert, D. Isochorismate pyruvate lyase: a pericyclic reaction mechanism? J. Am. Chem. Soc. 127, 15002–15003 (2005).
Tian, Z. et al. An enzymatic [4+2] cyclization cascade creates the pentacyclic core of pyrroindomycins. Nat. Chem. Biol. 11, 259–265 (2015).
Woodward, R. B. & Hoffmann, R. The conservation of orbital symmetry. Angew. Chem. Int. Ed. 8, 781–853 (1969).
Nicolaou, K. C., Snyder, S. A., Montagnon, T. & Vassilikogiannakis, G. The Diels–Alder reaction in total synthesis. Angew. Chem. Int. Ed. 41, 1668–1698 (2002).
Corey, E. J. & Cheng, X. M. The Logic of Chemical Synthesis (Wiley, 1989).
Hoffmann, H. M. R. The ene reaction. Angew. Chem. Int. Ed. 8, 556–577 (1969).
Alder, K. in Nobel Lectures, Chemistry 1942–1962 267–303 (Elsevier, 1964).
Niu, D. & Hoye, T. R. The aromatic ene reaction. Nat. Chem. 6, 34–40 (2014).
Mikami, K. & Shimizu, M. Asymmetric ene reactions in organic synthesis. Chem. Rev. 92, 1021–1050 (1992).
Jensen, A. W., Mohanty, D. K. & Dilling, W. L. The growing relevance of biological ene reactions. Bioorg. Med. Chem. 27, 686–691 (2019).
Lin, C.-I., McCarty, R. M. & Liu, H. The enzymology of organic transformations: a survey of name reactions in biological systems. Angew. Chem. Int. Ed. 56, 3446–3489 (2017).
Snider, B. B. & Lu, Q. Total synthesis of (±)-pyridoxatin. J. Org. Chem. 59, 8065–8070 (1994).
Snider, B. B. & Qing, L. A two-step synthesis of pyridoxatin analogues. Tetrahedr. Lett. 35, 531–534 (1994).
Jones, I. L., Moore, F. K. & Chai, C. L. L. Total synthesis of (±)-cordypyridones A and B and related epimers. Org. Lett. 11, 5526–5529 (2009).
Cai, Y. et al. Structural basis for stereoselective dehydration and hydrogen-bonding catalysis by the SAM-dependent pericyclase LepI. Nat. Chem. 11, 812–820 (2019).
Appendino, G., Cravotto, G., Toma, L., Annunziata, R. & Palmisano, G. The chemistry of coumarin derivatives. Part VI. Diels-Alder trapping of 3-methylene-2,4-chromandione. A new entry to substituted pyrano[3,2-c]coumarins. J. Org. Chem. 59, 5556–5564 (1994).
Qiao, Y. et al. Asperpyridone A: an unusual pyridone alkaloid exerts hypoglycemic activity through the insulin signaling pathway. J. Nat. Prod. 82, 2925–2930 (2019).
McBrien, K. D. et al. Fusaricide, a new cytotoxic N-hydroxypyridone from Fusarium sp. J. Nat. Prod. 59, 1151–1153 (1996).
Li, C., Sarotti, A. M., Yang, B., Turkson, J. & Cao, S. A new N-methoxypyridone from the co-cultivation of hawaiian endophytic fungi Camporesia sambuci FT1061 and Epicoccum sorghinum FT1062. Molecules 22, 1166 (2017).
Lee, H. J. et al. Pyridoxatin, an inhibitor of gelatinase A with cytotoxic activity. J. Microbiol. Biotechnol. 6, 445–450 (1996).
Singh, M. S., Nagaraju, A., Anand, N. & Chowdhury, S. Ortho-quinone methide (o-QM): a highly reactive, ephemeral and versatile intermediate in organic synthesis. RSC Advances 4, 55924–55959 (2014).
Singh, S. et al. Structural characterization of the mitomycin 7-O-methyltransferase. Proteins Struct. Funct. Bioinformat. 79, 2181–2188 (2011).
Kabsch, W. XDS. Acta Crystallogr. D Biol. Crystallogr. 66, 125–132 (2010).
Minor, W., Cymboriwski, M., Otwinowski, Z. & Chruszcz, M. HKL-3000: the integration ofdata reduction and structure solution-from diffraction images to an initial model in minutes. Acta Crystallogr. D Biol. Crystallogr. 62, 859–866 (2006).
Adams, P. D. et al. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. D Biol. Crystallogr. 66, 213–221 (2010).
McCoy, A. J. et al. Phaser crystallographic software. J. Appl. Cryst. 40, 658–674 (2007).
Emsley, P., Lohkamp, B., Scott, W. G. & Cowtan, K. Features and development of Coot. Acta Crystallogr. D Biol. Crystallogr. 66, 486–501 (2010).
Laskowski, R. A., MacArthur, M. W., Moss, D. S. & Thornton, J. M. ProCheck: a program to check the stereochemical quality of protein structures. J. Appl. Cryst. 26, 283–291 (1993).
Davis, I. W. et al. MolProbity: all-atom contacts and structure validation for proteins and nucleic acids. Nucleic Acids Res. 35, W375–W383 (2007).
DeLano, W. L. PyMOL: an open-source molecular graphics tool. Ccp4 Newslett. Protein Crystallogr 40, 11 (2002).
Schrödinger Release 2017-2: MacroModel, version 11.2.014 (Schrödinger, 2017).
Frisch, M. J. et al. Gaussian 16 Revision A.03 (Gaussian, 2016).
Chai, J.-D. & Head-Gordon, M. Long-range corrected hybrid density functionals with damped atom–atom dispersion corrections. Phys. Chem. Chem. Phys. 10, 6615–6620 (2008).
Krishnan, R., Binkley, J. S., Seeger, R. & Pople, J. A. Self-consistent molecular orbital methods. XX. A basis set for correlated wave functions. J. Chem. Phys. 72, 650–654 (1980).
Rassolov, V. A., Pople, J. A., Ratner, M. A. & Windus, T. L. 6-31G* basis set for atoms K through Zn. J. Chem. Phys. 109, 1223–1229 (1998).
Francl, M. M. et al. Self-consistent molecular orbital methods. XXIII. A polarization-type basis set for second-row elements. J. Chem. Phys. 77, 3654–3665 (1982).
Dill, J. D. & Pople, J. A. Self-consistent molecular orbital methods. XV. Extended Gaussian-type basis sets for lithium, beryllium, and boron. J. Chem. Phys. 62, 2921–2923 (1975).
Hehre, W. J., Ditchfield, R. & Pople, J. A. Self-consistent molecular orbital methods. XII. further extensions of Gaussian-type basis sets for use in molecular orbital studies of organic molecules. J. Chem. Phys. 56, 2257–2261 (1972).
Linder, M. & Brinck, T. On the method-dependence of transition state asynchronicity in Diels–Alder reactions. Phys. Chem. Chem. Phys. 15, 5108–5114 (2013).
Mardirossian, N. & Head-Gordon, M. Thirty years of density functional theory in computational chemistry: an overview and extensive assessment of 200 density functionals. Mol. Phys. 115, 2315–2372 (2017).
Clark, T., Chandrasekhar, J., Spitznagel, G. W. & Schleyer, P. V. R. Efficient diffuse function-augmented basis sets for anion calculations. III. The 3-21+G basis set for first-row elements, Li–F. J. Comput. Chem. 4, 294–301 (1983).
Blaudeau, J.-P., McGrath, M. P., Curtiss, L. A. & Radom, L. Extension of Gaussian-2 (G2) theory to molecules containing third-row atoms K and Ca. J. Chem. Phys. 107, 5016–5021 (1997).
Schäfer, A., Huber, C. & Ahlrichs, R. Fully optimized contracted Gaussian basis sets of triple zeta valence quality for atoms Li to Kr. J. Chem. Phys. 100, 5829–5835 (1994).
Weigend, F. & Ahlrichs, R. Balanced basis sets of split valence, triple zeta valence and quadruple zeta valence quality for H to Rn: Design and assessment of accuracy. Phys. Chem. Chem. Phys. 7, 3297–3305 (2005).
Case, D. A. et al. AMBER 2016 (UCSF, 2016).
Maier, J. A. et al. ff14SB: improving the accuracy of protein side chain and backbone parameters from ff99SB. J. Chem. Theory Comput. 11, 3696–3713 (2015).
Acknowledgements
This work was supported by grants from the NIH (1R01AI141481) to Y.T., K.N.H., and N.K.G., the NSF (CHE-1806581) to Y.T. and K.N.H., and the NSFC (91856202) and SMSTC (18430721500, 19XD1404800) to J.Z. C.S.J. is supported by generous funding through the Saul Winstein Fellowship. The authors thank the staff of beamlines BL17U1, BL18U1 and BL19U1 of Shanghai Synchrotron Radiation Facility for access and help with the X-ray data collection. Y.C. thanks J. Gan for assistance with structure refinement.
Author information
Authors and Affiliations
Contributions
M.O., C.S.J., K.N.H. and Y.T. developed the hypothesis and conceived the idea for the study. M.O., C.S.J., Y.C., J.Z., K.N.H. and Y.T. designed the experiments. M.O. performed all in vivo and in vitro experiments, as well as compound isolation and characterization. D.T., D.K., S.C. and M.-C.T. performed compound isolation and characterization. S.M.A., J.V.C., J.S.B., E.P. and N.K.G designed and performed synthesis of compounds. M.O. and T.B.K. performed bioinformatic analysis to identify the biosynthetic gene cluster. M.O. and Y.C. performed protein purification. Y.C. preformed all structural biology. C.S.J. performed all computational experiments. All authors analysed and discussed the results. M.O., C.S.J., Y.C., J.Z., K.N.H. and Y.T. prepared the manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Additional information
Peer review information Nature thanks Satish Nair and the other, anonymous, reviewer(s) for their contribution to the peer review of this work.
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data figures and tables
Extended Data Fig. 1 Density functional theory calculations for non-enzymatic Alder-ene and hetero-Diels–Alder reactions from (Z)-QM and (E)-QM and Alder-ene theozyme.
a, Transition states, products and energies for eight hetero-Diels–Alder and Alder-ene reactions are shown. In the transition states, the Alder-ene reactions adopt a conformation where the pyridone and forming cyclohexane are perpendicular to each other compared to the hetero-Diels–Alder reactions that are more co-planar in geometry. The Alder-ene reactions are synchronous and the hetero-Diels–Alder reactions are asynchronous, but concerted. TS-1 and TS-2 lead to 8’ and 9 with barriers of 25.1 and 23.2 kcal·mol–1, respectively. The structures 8a’, b’, c’, 9b, c and 11 are isomers of natural product scaffolds with barriers greater than TS-1. b, Alder-ene theozyme of (Z)-quinone methide complex leading to Alder-ene adduct (8’), O4- and O2-hetero-Diels–Alder adducts (9 and 11) with energies reported as enthalpies and Gibbs free energies.
Extended Data Fig. 2 Homologous biosynthetic gene clusters of pyridoxatin (1) and fusaricide (3) and the functions of PdxI and EpiI homologues.
a, Putative biosynthetic gene clusters of 1 and 3 (and 2), and their homologous biosynthetic gene clusters found in NCBI database. b, Key active site residues shown in an alignment with those from PdxI and EpiI homologues. Key residues involved in PdxI and EpiI catalysis are colored. c, In vitro analysis of PdxG and selected pericyclases using 5 as the starting substrate. The periselectivity can be correlated with the identity of the amino acid at position 413 (in PdxI, indicated in red dashed box). If valine occupies this position, the enzyme catalyses the Alder-ene reaction. On the other hand, if methionine occupies the position, the enzyme catalyses the hetero-Diels–Alder reaction.
Extended Data Fig. 3 Biochemical characterization of the ketoreductase PdxG.
a, In vitro reaction of 3 μM PdxG with 2 mM NADPH using 600 μM 5 as the substrate. 60% conversion from 5 to 6 was observed within 20 min. b, Kinetic analysis of PdxG-catalysed reduction of 5. Reaction mixtures containing 3 μM PdxG, 2 mM NADPH and different concentrations of 5 (10 μM to 1.2 mM) were incubated at 30 °C for 20 min. Error bars indicate s.d. of three independent replicates. c, Formation of 10 from 6 in the presence of 2 mM NADPH can be observed both with and without PdxG. Compound 6 was obtained from chemical reduction of 5 with NaBH4. Since 10 can be formed in the presence of only NADPH, we conclude NADPH can nonenzymatically reduce the QM to 10, which accounts for the result in Fig. 1f.
Extended Data Fig. 4 Biochemical characterization of PdxI and EpiI.
a, LC/MS analyses of chemically denatured PdxI and EpiI show no trace of SAM after purification. b, HPLC analyses of in vitro reaction of 150 μM 5 with 3 μM PdxG, 1 mM NADPH and 30 μM PdxI or 20 μM EpiI at 30 °C for 2 h in the presence or absence of cofactors. SAM or SAH does not alter the enzymatic activity of PdxI and EpiI. c, Since the interconversion of 8 and 9 could be envisioned by hydroalkoxylation/retro-hydroalkoxylation, we examined the possibility that PdxI and EpiI could catalyse the reaction of 9 to 8 and 8 to 9, respectively. The in vitro reactions of 100 μM 9 or 8 with 50 μM PdxI or 40 μM EpiI at 30 °C for 24 h were performed. However, no conversion of 9 to 8 and 8 to 9 was observed. d, In vitro reaction of PdxI and EpiI using 6 as the substrate. To obtain 6 for in vitro reaction, we chemically reduced 5 by NaBH4. Since this reduction proceeds non-stereoselectively, 6 and diastereomer 6′ were formed. After isolation of 6 and 6′ by HPLC, fractions containing 6 were not concentrated because of the instability and were immediately used as the substrate for PdxI and EpiI. e, LC/MS analysis of in vitro reactions catalysed by pericyclases using 6 as the substrate. Shown are compounds detected by selected ion monitoring at (M+H)+ of 248. In this mode, 6 is detected as the fragment ion. In the absence of either enzyme, 6 was converted to several products nonenzymatically, including 11. Minor compounds not isolated are indicated with *. In the presence of PdxI, 6 was nearly all converted to 8. In the presence of EpiI, 6 was nearly all converted to 9. Mutation of T232S in PdxI or T231A in EpiI changed selectivities of the enzymes to give other products, including 11. For additional mutagenesis data, please see Extended Data Figs. 7, 8.
Extended Data Fig. 5 Overlays of crystal structures with transition state structures for Alder-ene and hetero-Diels–Alder reactions.
a, Overlay of Alder-ene TS-3 with 5 bound in PdxI. Note the extended conformation of the alkyl chain versus the folded transition state geometry. The pyridone is bound by hydrogen bonds from K337, H161, Q412, and water mediated hydrogen bonds from T232, D233 and H336. b, Overlay O4-hetero-Diels–Alder TS-6 with 5 bound in HpiI. The pyridone is bound by hydrogen bonds from H161, Q414, and water mediated hydrogen bonds from T232, D233 and H338. Note that the K339 hydrogen bond to the pyridone O4 is not present in this structure. c, Overlay of PdxI-5 and HpiI-5. Omit maps not shown for clarity. Both PdxI and HpiI bind the pyridone such that it is prone to a syn-dehydration assisted by K337 (PdxI) or water molecule W (HpiI) and water molecules surrounding C7. The inset shows how V413 (in PdxI) or M415 (in HpiI) affects the orientation of the lysine residue (K337 in PdxI or K339 in HpiI) and its ability to hydrogen bond to the 4-OH of the pyridone. d, Overlay of PdxI-5, HpiI-5, Alder-ene TS-3, O4-hetero-Diels–Alder TS-6 and O2-hetero-Diels–Alder TS-5. Omit maps not shown for clarity. TS-3 and TS-6 bind in the active site sans disfavourable interactions whereas TS-5 clashes with T232. As both the calculated Alder-ene transition structure TS-3 and hetero-Diels–Alder TS-6 are quite similar in geometry and both easily fit into the PdxI active site, PdxI cannot solely rely on shape complementarity to catalyse the reaction with observed periselectivity. e, Chain B active site of PdxI-product (8) complex. Note the closer distances between K337 and the pyridone O4, the change in coordination of water mediated hydrogen bond from H336, and H161 shifting from a N1 hydrogen bond (in PdxI-5) to an O2 hydrogen bond. f, Overlay of Alder-ene TS-3 with 8 bound in PdxI. Note high similarity in structures of 8 and TS-3. This suggests that the enzyme distorts the product structure towards that of the Alder-ene transition state.
Extended Data Fig. 6 Molecular dynamic simulation of 7 in the PdxI active site.
Distances over time of hydrogen bonds to the various positions of the pyridone are tracked in chain A (a) and chain B (b) of the active site. Left panels show H336 and K337 form hydrogen bonds to the 4-position substituent on the pyridone ring. Right panel shows Q412 and H161 remain hydrogen bonded to 2-position substituent and pyridone nitrogen N1, respectively, for the majority of the simulation. c, Molecular dynamic simulations were initiated from an extended conformation (dihedral = ~180°). Over time, we monitored this conformation to see if the alkyl chain could spontaneously fold to a reactive conformation (dihedral = –20°). Indeed, for short durations of the simulations we observe the chain folding into a reactive conformation for a pericyclic reaction.
Extended Data Fig. 7 HPLC analysis of in vitro reaction of PdxI and mutants.
Mutation of the catalytic base K337A abolished the activity, while K337R mutant could retain approximately 10% activity. Individual substitution of H336A, Q412A and H161A all completely abolished the activity. Mutation of D233A or D233N completely abolish enzymatic activity. In contrast, in the D233E mutant 60% of activity and the original periselectivity were retained. This suggests that the carboxylate group of D233 in PdxI is important for enzyme function. A single mutation, V413M is sufficient to change the periselectivity from Alder-ene (>98:2, 8:9) to hetero-Diels–Alder reaction (40:60, 8:9). Further, mutation of T232 to either alanine or serine, but not valine, can lead to the formation of the O2-hetero-Diels–Alder product 11 along with the Alder-ene product 8. The data show one representative experiment from at least three independent replicates. Reaction conditions: 150 μM 5 with 3 μM PdxG, 1 mM NADPH and 50 μM PdxI (wild type or mutant) at 30 °C for 2 h.
Extended Data Fig. 8 HPLC analysis of in vitro reaction of EpiI and mutants.
In contrast to PdxI, substitution of K338 to alanine did not abolished and retained the activity (83%) (Extended Data Fig. 9b). H336A, H161A and Q410A (corresponding to Q412 in PdxI) mutants were highly insoluble and cannot be assayed. Although D232A and D232N mutations completely abolished the enzymatic activity, the D233E mutation retained 53% of activity and maintained the original periselectivity. This suggests that the carboxylate group of D232 in EpiI is also important for enzyme function. Mutation of Y205F retained 89% activity and maintained the original periselectivity, suggesting the hydroxy group of Y205 is not essential for catalysis. The M411V (corresponding to V413 in PdxI) and M411C mutations increased the Alder-ene product ratio compared to the wild type of EpiI. The T232A and T232S mutations but not T232V mutation, generated the O2-hetero-Diels–Alder product 11 and the Alder-ene product 8 as the minor products, with the hetero-Diels–Alder product 9 as a major product. The double mutation M411V/T231A of EpiI reversed the periselectivity from the native hetero-Diels–Alder reaction (<5:95, 8:9) to the energetically disfavoured Alder-ene reaction (2:1, 8:9), although the enzymatic activity is only moderately decreased (Extended Data Fig. 9). In the double mutant, 11 was formed due to the mutation of T231. Other double mutants such as M411V/T231S, M411T/T231A, M411C/T231A and M411G/T231A also reversed periselectivity. The data shown are that of one representative experiment from at least three independent replicates. Reaction condition: 300 μM 5 with 3 μM PdxG, 1 mM NADPH and 40 μM EpiI (wild type or mutant) at 30 °C for 2 h.
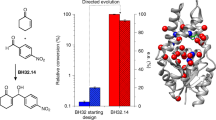
Extended Data Fig. 9 Relative activities of PdxI, EpiI and mutants.
The activity of each mutant is compared to that of wild-type PdxI or EpiI quantified by the formation of 8, 9 and 11. Error bars indicate s.d. of three independent replicates. Asterisks indicate mutants with no measurable activity. a, The relative enzymatic activity of PdxI mutants. Reaction conditions: 150 μM 5 with 3 μM PdxG, 1 mM NADPH and 50 μM PdxI mutants at 30 °C for 2 h. b, The relative activity of EpiI mutants. Reaction condition: 300 μM 5 with 3 μM PdxG, 1 mM NADPH and 40 μM EpiI mutants at 30 °C for 2 h.
Extended Data Fig. 10 Proposed mechanisms of PdxI- and EpiI-catalysed reactions.
a, The catalytic cycle of PdxI-catalysed reaction is initiated by the deprotonation of the 4-hydroxy group by K337 followed by the syn-dehydration to 7 assisted by the extend water hydrogen bonding network mediated by H336. Subsequently, protonated K337 serves as the general acid catalyst and forms the strong hydrogen bonding with 4-carbonyl oxygen of 7 to set the stage for the periselective Alder-ene reaction. Note that the steric effect of T232 inhibits the formation of the O2-hetero-Diels–Alder product 11 to further control regioselectivity. The alkyl chain folds to a reactive conformation and readily undergoes an Alder-ene reaction. After this, the tautomerization is facilitated by K337 and possibly water mediated by H336 to form and release 8. Then, the next catalytic cycle initiates. b, The catalytic cycle of EpiI-catalysed reaction, in contrast to PdxI, is initiated by the deprotonation of the hydroxy group by an alternative general base, possibly water followed by the syn-dehydration to 7. Since the key lysine residue does not form hydrogen bonding with 4-carbonyl oxygen of 7 due to the bulkier side chain of M411 (corresponding to V413 in PdxI), the favoured hetero-Diels–Alder reaction takes place to form and release 9. As same as PdxI, the steric effect of T231 inhibits the formation of the O2-hetero-Diels–Alder product 11 to further control regioselectivity. Then, the next catalytic cycle initiates.
Supplementary information
Supplementary Information
This file contains Supplementary Methods, Supplementary Tables S1-S14 and Supplementary Notes 1-10.
Rights and permissions
About this article
Cite this article
Ohashi, M., Jamieson, C.S., Cai, Y. et al. An enzymatic Alder-ene reaction. Nature 586, 64–69 (2020). https://doi.org/10.1038/s41586-020-2743-5
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41586-020-2743-5
This article is cited by
-
O-methyltransferase-like enzyme catalyzed diazo installation in polyketide biosynthesis
Nature Communications (2023)
-
Enzymatic catalysis favours eight-membered over five-membered ring closure in bicyclomycin biosynthesis
Nature Catalysis (2023)
-
Tandem intermolecular [4 + 2] cycloadditions are catalysed by glycosylated enzymes for natural product biosynthesis
Nature Chemistry (2023)
-
Discovery and characterization of a terpene biosynthetic pathway featuring a norbornene-forming Diels-Alderase
Nature Communications (2022)
-
Discovery and investigation of natural Diels–Alderases
Journal of Natural Medicines (2021)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.