Abstract
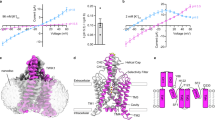
TASK2 (also known as KCNK5) channels generate pH-gated leak-type K+ currents to control cellular electrical excitability1,2,3. TASK2 is involved in the regulation of breathing by chemosensory neurons of the retrotrapezoid nucleus in the brainstem4,5,6 and pH homeostasis by kidney proximal tubule cells7,8. These roles depend on channel activation by intracellular and extracellular alkalization3,8,9, but the mechanistic basis for TASK2 gating by pH is unknown. Here we present cryo-electron microscopy structures of Mus musculus TASK2 in lipid nanodiscs in open and closed conformations. We identify two gates, distinct from previously observed K+ channel gates, controlled by stimuli on either side of the membrane. Intracellular gating involves lysine protonation on inner helices and the formation of a protein seal between the cytoplasm and the channel. Extracellular gating involves arginine protonation on the channel surface and correlated conformational changes that displace the K+-selectivity filter to render it nonconductive. These results explain how internal and external protons control intracellular and selectivity filter gates to modulate TASK2 activity.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
Data availability
The TASK2 protein sequence is available from Uniprot accession Q9JK62. The final maps of TASK2 in MSP1D1 nanodiscs at pH 8.5 and pH 6.5 have been deposited to the Electron Microscopy Data Bank under accession codes 21846 and 21843. Atomic coordinates have been deposited in the Protein Data Bank under IDs 6WM0 and 6WLV. Original micrograph movies have been deposited to EMPIAR under accession codes EMPIAR-10422 and EMPIAR-10423.
References
Reyes, R. et al. Cloning and expression of a novel pH-sensitive two pore domain K+ channel from human kidney. J. Biol. Chem. 273, 30863–30869 (1998).
Enyedi, P. & Czirják, G. Molecular background of leak K+ currents: two-pore domain potassium channels. Physiol. Rev. 90, 559–605 (2010).
Cid, L. P. et al. TASK-2: a K2P K+ channel with complex regulation and diverse physiological functions. Front. Physiol. 4, 198 (2013).
Wang, S. et al. TASK-2 channels contribute to pH sensitivity of retrotrapezoid nucleus chemoreceptor neurons. J. Neurosci. 33, 16033–16044 (2013).
Guyenet, P. G. et al. The retrotrapezoid nucleus: central chemoreceptor and regulator of breathing automaticity. Trends Neurosci. 42, 807–824 (2019).
Gestreau, C. et al. Task2 potassium channels set central respiratory CO2 and O2 sensitivity. Proc. Natl Acad. Sci. USA 107, 2325–2330 (2010).
Warth, R. et al. Proximal renal tubular acidosis in TASK2 K+ channel-deficient mice reveals a mechanism for stabilizing bicarbonate transport. Proc. Natl Acad. Sci. USA 101, 8215–8220 (2004).
López-Cayuqueo, K. I., Peña-Münzenmayer, G., Niemeyer, M. I., Sepúlveda, F. V. & Cid, L. P. TASK-2 K2P K+ channel: thoughts about gating and its fitness to physiological function. Pflugers Arch. 467, 1043–1053 (2015).
Niemeyer, M. I., Cid, L. P., Peña-Münzenmayer, G. & Sepúlveda, F. V. Separate gating mechanisms mediate the regulation of K2P potassium channel TASK-2 by intra- and extracellular pH. J. Biol. Chem. 285, 16467–16475 (2010).
Niemeyer, M. I., Cid, L. P., Barros, L. F. & Sepúlveda, F. V. Modulation of the two-pore domain acid-sensitive K+ channel TASK-2 (KCNK5) by changes in cell volume. J. Biol. Chem. 276, 43166–43174 (2001).
Bayliss, D. A., Barhanin, J., Gestreau, C. & Guyenet, P. G. The role of pH-sensitive TASK channels in central respiratory chemoreception. Pflugers Arch. 467, 917–929 (2015).
Julio-Kalajzić, F. et al. K2P TASK-2 and KCNQ1–KCNE3 K+ channels are major players contributing to intestinal anion and fluid secretion. J. Physiol. 596, 393–407 (2018).
Clark, R. B., Kondo, C., Belke, D. D. & Giles, W. R. Two-pore domain K+ channels regulate membrane potential of isolated human articular chondrocytes. J. Physiol. 589, 5071–5089 (2011).
Alvarez-Baron, C. P., Jonsson, P., Thomas, C., Dryer, S. E. & Williams, C. The two-pore domain potassium channel KCNK5: induction by estrogen receptor alpha and role in proliferation of breast cancer cells. Mol. Endocrinol. 25, 1326–1336 (2011).
Reed, A. P., Bucci, G., Abd-Wahab, F. & Tucker, S. J. Dominant-negative effect of a missense variant in the TASK-2 (KCNK5) K+ channel associated with Balkan endemic nephropathy. PLoS One 11, e0156456 (2016).
Kang, D. & Kim, D. Single-channel properties and pH sensitivity of two-pore domain K+ channels of the TALK family. Biochem. Biophys. Res. Commun. 315, 836–844 (2004).
Niemeyer, M. I. et al. Neutralization of a single arginine residue gates open a two-pore domain, alkali-activated K+ channel. Proc. Natl Acad. Sci. USA 104, 666–671 (2007).
Brohawn, S. G., Campbell, E. B. & MacKinnon, R. Physical mechanism for gating and mechanosensitivity of the human TRAAK K+ channel. Nature 516, 126–130 (2014).
Miller, A. N. & Long, S. B. Crystal structure of the human two-pore domain potassium channel K2P1. Science 335, 432–436 (2012).
Lolicato, M. et al. K2P2.1 (TREK-1)–activator complexes reveal a cryptic selectivity filter binding site. Nature 547, 364–368 (2017).
Dong, Y. Y. et al. K2P channel gating mechanisms revealed by structures of TREK-2 and a complex with Prozac. Science 347, 1256–1259 (2015).
Lolicato, M. et al. K2P channel C-type gating involves asymmetric selectivity filter order–disorder transitions. Preprint at https://doi.org/10.1101/2020.03.20.000893 (2020).
Rödström, K. E. J. et al. A lower X-gate in TASK channels traps inhibitors within the vestibule. Nature 582, 443–447 (2020).
Niemeyer, M. I., Cid, L. P., Paulais, M., Teulon, J. & Sepúlveda, F. V. Phosphatidylinositol (4,5)-bisphosphate dynamically regulates the K2P background K+ channel TASK-2. Sci. Rep. 7, 45407–45414 (2017).
Ritchie, T. K. et al. in Methods in Enzymology Vol. 464 (ed. Düzgünes, N.) 211–231 (2009).
Isom, D. G., Castañeda, C. A., Cannon, B. R. & García-Moreno, B. Large shifts in pKa values of lysine residues buried inside a protein. Proc. Natl Acad. Sci. USA 108, 5260–5265 (2011).
del Camino, D. & Yellen, G. Tight steric closure at the intracellular activation gate of a voltage-gated K+ channel. Neuron 32, 649–656 (2001).
Jiang, Y. et al. The open pore conformation of potassium channels. Nature 417, 523–526 (2002).
Piechotta, P. L. et al. The pore structure and gating mechanism of K2P channels. EMBO J. 30, 3607–3619 (2011).
Schewe, M. et al. A non-canonical voltage-sensing mechanism controls gating in K2P K+ channels. Cell 164, 937–949 (2016).
Bagriantsev, S. N., Peyronnet, R., Clark, K. A., Honoré, E. & Minor, D. L. Jr. Multiple modalities converge on a common gate to control K2P channel function. EMBO J. 30, 3594–3606 (2011).
Hoshi, T., Zagotta, W. N. & Aldrich, R. W. Two types of inactivation in Shaker K+ channels: effects of alterations in the carboxy-terminal region. Neuron 7, 547–556 (1991).
Cohen, A., Ben-Abu, Y., Hen, S. & Zilberberg, N. A novel mechanism for human K2P2.1 channel gating. Facilitation of C-type gating by protonation of extracellular histidine residues. J. Biol. Chem. 283, 19448–19455 (2008).
Cuello, L. G., Cortes, D. M. & Perozo, E. The gating cycle of a K+ channel at atomic resolution. eLife 6, e28032 (2017).
Pau, V., Zhou, Y., Ramu, Y., Xu, Y. & Lu, Z. Crystal structure of an inactivated mutant mammalian voltage-gated K+ channel. Nat. Struct. Mol. Biol. 24, 857–865 (2017).
Brohawn, S. G. How ion channels sense mechanical force: insights from mechanosensitive K2P channels TRAAK, TREK1, and TREK2. Ann. NY Acad. Sci. 1352, 20–32 (2015).
Brohawn, S. G. et al. The mechanosensitive ion channel TRAAK is localized to the mammalian node of Ranvier. eLife 8, e50403 (2019).
Mastronarde, D. N. Automated electron microscope tomography using robust prediction of specimen movements. J. Struct. Biol. 152, 36–51 (2005).
Zivanov, J., Nakane, T. & Scheres, S. H. W. A Bayesian approach to beam-induced motion correction in cryo-EM single-particle analysis. IUCrJ 6, 5–17 (2019).
Zheng, S. Q. et al. MotionCor2: anisotropic correction of beam-induced motion for improved cryo-electron microscopy. Nat. Methods 14, 331–332 (2017).
Zivanov, J. et al. New tools for automated high-resolution cryo-EM structure determination in RELION-3. eLife 7, e42166 (2018).
Rohou, A. & Grigorieff, N. CTFFIND4: fast and accurate defocus estimation from electron micrographs. J. Struct. Biol. 192, 216–221 (2015).
Punjani, A., Rubinstein, J. L., Fleet, D. J. & Brubaker, M. A. cryoSPARC: algorithms for rapid unsupervised cryo-EM structure determination. Nat. Methods 14, 290–296 (2017).
Afonine, P. V. et al. Real-space refinement in PHENIX for cryo-EM and crystallography. Acta Crystallogr. D 74, 531–544 (2018).
Ramírez-Aportela, E. et al. Automatic local resolution-based sharpening of cryo-EM maps. Bioinformatics 36, 765–772 (2020).
Chen, V. B. et al. MolProbity: all-atom structure validation for macromolecular crystallography. Acta Crystallogr. D 66, 12–21 (2010).
Emsley, P., Lohkamp, B., Scott, W. G. & Cowtan, K. Features and development of Coot. Acta Crystallogr. D 66, 486–501 (2010).
Acknowledgements
We thank J. Remis, D. Toso and P. Tobias at UC Berkeley for assistance with microscope set-up and data collection; Z. Fu and N. Kostow for help with initial cloning and screening; and members of the Brohawn laboratory for feedback on the manuscript. S.G.B. is a New York Stem Cell Foundation–Robertson Neuroscience Investigator. This work was supported by the New York Stem Cell Foundation, NIGMS grant DP2GM123496, a McKnight Foundation Scholar Award, a Klingenstein-Simons Foundation Fellowship Award and a Sloan Research Fellowship to S.G.B.
Author information
Authors and Affiliations
Contributions
B.L., R.A.R. and S.G.B. conceived the project. R.A.R. generated and screened the constructs. R.A.R. optimized protein expression and purification. B.L. performed protein purification and nanodisc reconstitution for cryo-EM. B.L. prepared samples for cryo-EM, collected cryo-EM data and processed cryo-EM data. B.L. built and refined the atomic models. B.L., R.A.R. and S.G.B. performed electrophysiology. B.L., R.A.R. and S.G.B. wrote the manuscript. S.G.B. supervised the project and secured funding.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Peer review information Nature thanks Douglas Bayliss, Francisco Sepúlveda and the other, anonymous, reviewer(s) for their contribution to the peer review of this work.
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data figures and tables
Extended Data Fig. 1 Purification and reconstitution of TASK2.
a–d, Data for assembly of TASK2-nanodisc samples at pH 8.5. a, Chromatogram from a Superdex 200 gel filtration of TASK2 purified in DDM/CHS. b, Coomassie-stained SDS–PAGE of pooled TASK2-containing fractions indicated by grey bar in a. c, Chromatogram from Superdex 200 gel filtration of TASK2 reconstituted in MSP1D1 lipid nanodiscs. d, Coomassie-stained SDS–PAGE of final pooled TASK2-MSP1D1 nanodisc sample indicated by grey bar in c. e–h, Same as a–d, but for samples at pH 6.5. Purifications were performed once. Gels were run once. For gel source data, see Supplementary Fig. 3.
Extended Data Fig. 2 pH and PtdIns(4,5)P2 dependence of TASK2 and pH dependence of TASK2 mutant constructs.
a–d, pHint (a, b) and pHext (c, d) dependence from a representative cell expressing full-length TASK2 (a, c) and the C-terminally truncated TASK2 construct used for structure determination (b, d). Normalized fold-activation of current by alkaline pHint (pHint = 9/pHint = 7 at 0 mV) (a, b) or alkaline pHext (c, d) versus pH is plotted. Mean ± s.e.m. from three sweeps are plotted. Fits to Hill equations are drawn with pK1/2 = 7.7, 7.9, 7.8, and 7.6 and Hill slope = 1.0, 1.1, 1.1, and 1.2 for a–d, respectively. Plots are shown with different scales. e, Current-voltage relationships from a representative inside-out patch from a TASK2-expressing cell before (circles) and after (squares) the addition of 50μM C8-PtdIns(4,5)P2. f–j, Current–voltage relationships recorded from a representative cell expressing TASK2 mutants K245A, N243A, N243R, W244A, and N243K/K245N with pHint = 7 (circles) and pHint = 9 (squares). k–q, Current–voltage relationships recorded from a representative cell expressing TASK2 mutants K245A, R224A, V104A, N87A, N87S, N82A, and E228A with pHext = 7 (circles) and pHext = 9. (squares). Data in e–q are mean currents ± s.e.m. from three consecutive sweeps at the indicated voltage.
Extended Data Fig. 3 Cryo-EM processing pipeline for TASK2 at pH 8.5.
a, Example micrograph (left) and selected 2D class averages (right) of TASK2 in MSP1D1 lipid nanodiscs at pH 8.5. 2D classification was performed with an extracted box size of 200 pixels. b, Cryo-EM data processing steps in RELION and cryoSPARC2. See Methods for details. Cryo-EM data were collected once. The micrograph in a is representative of the 2,814 micrographs selected for analysis.
Extended Data Fig. 4 Cryo-EM processing pipeline for TASK2 at pH 6.5.
a, Example micrograph (left) and selected 2D class averages (right) of TASK2 in MSP1D1 lipid nanodiscs at pH 6.5. 2D classification was performed with an extracted box size of 160 pixels. b, Cryo-EM data processing steps in RELION and cryoSPARC2. See Methods for details. Cryo-EM data were collected once. The micrograph in a is representative of the 2,683 micrographs selected for analysis.
Extended Data Fig. 5 Cryo-EM validation.
a–h, Data for TASK2-nanodisc samples at pH 8.5 (a–d) and pH 6.5 (e–h). a, e, Angular distribution of particles used in final refinement with final map for reference. b, f, Local resolution estimated in RELION coloured as indicated on the final map. TM4s are indicated in a view from the cytoplasm. c, g, Fourier Shell Correlation (FSC) relationships between (black) the two unfiltered half-maps from refinement and used for calculating overall resolution at 0.143, (teal) final map versus model, (pink) half-map one versus model, and (orange) half-map two versus model. d, h, Cryo-EM density carved around TM4A and TM4B with the position of K245 indicated.
Extended Data Fig. 6 Extracellular ion pathways and lateral membrane openings in TASK2.
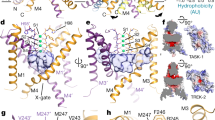
a, b, Structures of TASK2 determined at pH 8.5 (a) and pH 6.5 (b). The surface of a bifurcated extracellular pathway from the top of the selectivity filter underneath the helical cap to the extracellular solution on either side is shown in grey. c, Radius of the extracellular pathway as a function of distance from the conduction axis. The path is similarly accessible to K+ ions in both structures. d, e, View from the membrane plane of the cytoplasmic sides of TASK2 TM4 and TM2 from low pH (closed) (d) and high pH (open) (e) structures. Protein surface is shown half transparent. f, Change in channel cross sectional area upon opening as a function of membrane depth for TASK2 and TRAAK. TRAAK expands within the membrane upon opening while TASK2 constricts near the membrane-cytoplasm interface. g, h, View from the membrane plane of the cytoplasmic sides of TRAAK TM4 and TM2 from nonconductive (closed) (g) and conductive (open) (h) structures. i, Minimum cross-sectional areas of membrane-facing lateral openings in TASK2 closed, and TASK2 open, TRAAK closed, and TRAAK open structures. Cross-sectional areas for each structure correspond to the narrowest 1 Å segment of a path connecting the channel cavity and membrane bilayer calculated using a spherical probe. The cross-sectional area of a lipid acyl chain methylene group is drawn with a dashed line for comparison. An acyl chain could access the cavity of TASK2 and TRAAK channels in closed, but not open, conformations.
Extended Data Fig. 7 Comparison of selectivity filters and selectivity filter gates in TASK2 and other K+ channels.
a–n, Two opposing selectivity filter regions and positions of bound K+ ions from structures of the channels indicated: open TASK2 SF1 (a), closed TASK2 SF1 (b), overlaid open and closed TASK2 SF1s (c), open TASK2 SF2 (d), closed TASK2 SF2 (e), overlaid open and closed TASK2 SF2s (f), open Kv1.1-2.1 (g), inactivated Kv1.1-2.1 mutant V406W (h), overlaid open and inactivated Kv1.1-2.1 mutant V406W (i), overlaid inactivated Kv1.1-2.1 and closed TASK2 SF1 (j), open KcsA (k), low K+ inactivated KcsA (l), overlaid open and inactivated KcsA (m), and overlaid inactivated KcsA and closed TASK2 SF1 (n).
Extended Data Fig. 8 Comparison of inner gates in TASK2 and other K+ channels.
a–g, Stereo views from the membrane plane, highlighting inner gate regions of selected K+ channels: open MthK (a), closed KcsA (b), open TASK2 (c), closed TASK2 (d), and closed TASK1 (e). f, An overlay of the TASK2 open and closed structures with the canonical “bundle crossing” inner gating channels KcsA and MthK, coloured as in a–d. g, An overlay of the TASK2 open and closed structures with TASK1, with a distinct inner “X” gate, coloured as in c–e.
Supplementary information
Supplementary Figure
Supplementary Figure 1 | Sequence alignment of mouse K2P channels. Alignment of mouse K2P channels colored by conservation. TASK2 secondary structure is drawn above the sequences with gaps and unmodeled residues shown as dashed lines, loops and non-helical secondary structure as gray lines, and selectivity filters as green lines. Residues implicated in the extracellular gate are indicated with green boxes, residues implicated in the intracellular gate with red boxes, and disulfide bond forming C51 with a yellow box.
Supplementary Figure
Supplementary Figure 2 | Sequence alignment of TASK2 channels. Alignment of M. musculus, H. sapiens, G. gallus, X. laevis, D. rerio, M. domestica, and T. guttata TASK2 colored by conservation. TASK2 secondary structure is drawn above the sequences with gaps and unmodeled residues shown as dashed lines, loops and non-helical secondary structure as gray lines, and selectivity filters as green lines. Residues implicated in the extracellular gate are indicated with green boxes, residues implicated in the intracellular gate with red boxes, and disulfide bond forming C51 with a yellow box.
Supplementary Figure
Supplementary Figure 3 | SDS-PAGE gels of purified and reconstituted TASK2. Uncropped original gel images with regions shown in Extended Data Fig. 1 outlined in red. Molecular weight of bands in marker lane is indicated.
Rights and permissions
About this article
Cite this article
Li, B., Rietmeijer, R.A. & Brohawn, S.G. Structural basis for pH gating of the two-pore domain K+ channel TASK2. Nature 586, 457–462 (2020). https://doi.org/10.1038/s41586-020-2770-2
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41586-020-2770-2
This article is cited by
-
Conformational plasticity of NaK2K and TREK2 potassium channel selectivity filters
Nature Communications (2023)
-
Structural Basis for pH-gating of the K+ channel TWIK1 at the selectivity filter
Nature Communications (2022)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.