Abstract
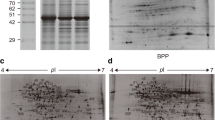
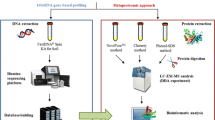
Soil protein extraction and database construction are the key points of soil proteomics research. In this paper, tea tree rhizosphere soil was used as material. The soil proteins were extracted three times by citrate, SDS, and mixture of citrate and SDS, respectively. The total proteins were separated by 2-DE electrophoresis and identified by Data Dependent Acquisition (DDA). The DDA data collection was further separated by High-Performance Liquid Chromatography (HPLC) and identified by LC–MS/MS, then to build the database of soil protein and microbial species using fungus and bacteria databases. The result showed soil protein was identified and reached 2741 points, and the molecular weight was mainly distributed in between 2.64 and 338.33 kDa, and Isoelectric point (pI) is mainly distributed in between 3.78 and 12.15. The soil protein was mainly from 138 families, 346 species of microorganisms. This optimization method could obtain more proteins than previous methods, with a wider range of molecular weight and pI. This study lays an important foundation for the research and development of soil metaproteomics.




Similar content being viewed by others
References
Abram F, Gunnigle E, O’Flaherty V (2009) Optimisation of protein extraction and 2-DE for metaproteomics of microbial communities from anaerobic wastewater treatment biofilms. Electrophoresis 30(23):4149–4151
Arafat Y, Wei X, Jiang Y, Chen T, Saqib H, Lin S, Lin W (2017) Spatial distribution patterns of root-associated bacterial communities mediated by root exudates in different aged ratooning tea monoculture systems. Int J Mol Sci 18(8):1727–1733
Chen SN, Rillig MC, Wang W (2009) Improving soil protein extraction for metaproteome analysis and glomalin-related soil protein detection. Proteomics 9(21):4970–4973
Chourey K, Jansson J, VerBerkmoes N, Shah M, Chavarria KL, Tom LM, Brodie EL, Hettich RL (2010) Direct cellular lysis protein extraction protocol for soil metaproteomics. J Proteome Res 9(12):6615–6622
Erickson BK, Mueller RS, VerBerkmoes NC, Shah M, Singer SW, Thelen MP, Banfield JF, Hettich RL (2010) Computational prediction and experimental validation of signal peptide cleavages in the extracellular proteome of a natural microbial community. J Proteome Res 9(5):2148–2159
Guo H, Nasir M, Lv J, Dai Y, Gao J (2017) Understanding the variation of microbial community in heavy metals contaminated soil using high throughput sequencing. Ecotoxicol Environ Safe 144:300–306
Harwkes C, Deangelis K, Firestore M (2007) Root interactions withsoil microbial communities and process. In: Cardon Z, Whitbeck J (eds) The rhizosphere: anecological perspective. Elsevier, Burlington, pp 1–25
Huang HJ, Chen WY, Wu JH (2014) Total protein extraction for metaproteomics analysis of methane producing biofilm: the effects of detergents. Int J Mol Sci 15:10169–10184
Lin R, Lin XM, Guo TT, Wu LK, Zhang WJ, Lin WX (2015) Metaproteomic analysis of bacterial communities in marine mudflat aquaculture sediment. World J Microbiol Biotechnol 31(9):1397–1408
Ling N, Deng KY, Song Y, Wu YC, Zhao J, Raza W, Huang QW, Shen Q (2014) Variation of rhizosphere bacterial community in watermelon continuous mono-cropping soil by long-term application of a novel bioorganic fertilizer. Microbiol Res 169:570–578
Li XG, Ding CF, Hua K, Zhang TL, Zhang YN, Zhao L, Yang YR, Liu JG, Wang XX (2014) Soil sickness of peanuts is attributable to modifications in soil microbes induced by peanut root exudates rather than to direct allelopathy. Soil Biol Biochem 78:149–159
Li YC, Li Z, Li ZW, Jiang YH, Weng BQ, Lin WX (2016) Variations of rhizosphere bacterial communities in tea (Camellia sinensis L) continuous cropping soil by high-throughput pyrosequencing approach. J Appl Microbiol 121(3):787–799
Mukerji K (2006) Rhizosphere biology. In: Mukerji KG, Manoharachary C, Singh J (eds) microbial activity in the rhizosphere. Springer, Berlin, pp 1–39
Nilsson RH, Anslan S, Bahram M, Wurzbacher C, Baldrian P, Tedersoo L (2019) Mycobiome diversity: high-throughput sequencing and identification of fungi. Nat Rev Microbiol 17(2):95–109
Paz-Ferreiro J, Fu SL (2016) Biological indices for soil quality evaluation: perspectives and limitations. Land Degrad Dev 27(1):14–25
Pierre-Alain M, Christophe M, Severine S, Houria A, Philippe L, Lionel R (2007) Protein extraction and fingerprinting optimization of bacterial communities in natural environment. Microb Ecol 53(3):426–434
Siggins A, Gunnigle E, Abram F (2012) Exploring mixed microbial community functioning: recent advances in metaproteomics. FEMS Microbiol Ecol 80(2):265–280
Stokke R, Roalkvam I, Lanzen A, Haflidason H, Steen IH (2012) Integrated metagenomic and metaproteomic analyses of an ANME-1-dominated community in marine cold seep sediments. Environ Microbiol 14(5):1333–1346
Utobo EB, Tewari L (2015) Soil enzymes as bioindicators of soil ecosystem status. Appl Ecol Environ Res 13(1):147–169
Vasconcellos RL, Bonfim JA, Baretta D, Cardoso EJ (2016) Arbuscular mycorrhizal fungi and glomalin-related soil protein as potential indicators of soil quality in a recuperation gradient of the Atlantic forest in Brazil. Land Degrad Dev 27(2):325–334
VerBerkmoes NC, Denef VJ, Hettich RL, Banfield JF (2009) Systems biology: functional analysis of natural microbial consortia using community proteomics. Nat Rev Microbiol 7(3):196–205
Wang HB, Zhang ZX, Li H, He HB, Fang CX, Zhang AJ, Li QS, Chen RS, Guo XK, Lin HF, Wu LK, Wu L, Wang H, Zhang Z, Lin R, Lin W (2011) Characterization of metaproteomics in crop rhizospheric soil. J Proteome Res 10:932–940
Wiśniewski JR, Zougman A, Nagaraj N, Mann M (2009) Universal sample preparation method for proteome analysis. Nat Methods 6(5):359–362
Wu LK, Wang HB, Zhang ZX, Lin R, Zhong ZY, Lin WX (2011) Comparative metaproteomic analysis on consecutively Rehmannia glutinosa monocultured rhizosphere soil. PLoS ONE 6(5):e20611
Wu LK, Li ZF, Li J, Khan MA, Huang WM, Zhang ZY, Lin WX (2013) Assessment of shifts in microbial community structure and catabolic diversity in response to Rehmannia glutinosa monoculture. Appl Soil Ecol 67:1–9
Wu LK, Huang WM, Wang JY, Wu HM, Chen J, Qin XJ, Zhang ZY, Lin WX (2015) Diversity analysis of rhizosphere microflora of wild R. glutinosa grown in monocropping for different years. Acta Agronomica Sinica 41(2):308–317 (in Chinese)
Yang YH, Chen DM, Jin Y, Wang HB, Duan YQ, Guo XK, He HB, Lin WX (2011) Effects of different fertilizers on function diversities of microbial flora in rhizospheric soil of monoculture tobacco. Acta Agronomica Sinica 37(1):105–111 (in Chinese)
Zhang W, Chen L, Zhang R, Lin K (2016) High throughput sequencing analysis of the joint effects of BDE209-Pb on soil bacterial community structure. J Hazard Mater 301:1–7
Zhou X, Wu F (2012) P-Coumaric acid influenced cucumber rhizosphere soil microbial communities and the growth of Fusarium oxysporum f. sp. cucumerinum Owen. PLoS ONE 7:e48288
Zhao YP, Wu LK, Chu LX, Yang YQ, Li ZF, Azeem S, Zhang ZX, Fang CX, Lin WX (2015) Interaction of Pseudostellaria heterophylla with Fusarium oxysporum f.sp. heterophylla mediated by its root exudates in a consecutive monoculture system. Sci Rep 5:8197
Acknowledgements
This work was supported by China Postdoctoral Science Foundation (2016M600493), the National 948 Project (2014-Z36), Natural Science Foundation of Fujian Province (2017J05057), Science and Technology Project of Longyan City (2017LY71), the Project of Scientific Research of Young and Middle-aged teachers, Fujian Province (JAT170573), Fujian Outstanding Research Talent Cultivation Project, National Program for Innovation and Entrepreneurship Training for College Students (201911312001, 202011312004), and Youth Top Talent Training Program of Longyan University (2019ZJ19).
Author information
Authors and Affiliations
Corresponding author
Additional information
Communicated by E. Schleiff.
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Wang, Hb., Zhu, Cl., Wang, Yh. et al. Protein extraction and database construction in tea rhizosphere soil. Acta Physiol Plant 42, 158 (2020). https://doi.org/10.1007/s11738-020-03146-5
Received:
Revised:
Accepted:
Published:
DOI: https://doi.org/10.1007/s11738-020-03146-5