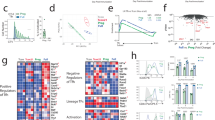
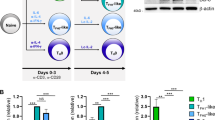
Abstract
T follicular helper (TFH) cells are critical in adaptive immune responses to pathogens and vaccines; however, what drives the initiation of their developmental program remains unclear. Studies suggest that a T cell antigen receptor (TCR)-dependent mechanism may be responsible for the earliest TFH cell–fate decision, but a critical aspect of the TCR has been overlooked: tonic TCR signaling. We hypothesized that tonic signaling influences early TFH cell development. Here, two murine TCR-transgenic CD4+ T cells, LLO56 and LLO118, which recognize the same antigenic peptide presented on major histocompatibility complex molecules but experience disparate strengths of tonic signaling, revealed low tonic signaling promotes TFH cell differentiation. Polyclonal T cells paralleled these findings, with naive Nur77 expression distinguishing TFH cell potential. Two mouse lines were also generated to both increase and decrease tonic signaling strength, directly establishing an inverse relationship between tonic signaling strength and TFH cell development. Our findings elucidate a central role for tonic TCR signaling in early TFH cell-lineage decisions.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout







Similar content being viewed by others
Data availability
The data that support the findings of this study are available from the corresponding author upon reasonable request.
References
Crotty, S. Follicular helper CD4 T cells (TFH). Annu. Rev. Immunol. 29, 621–663 (2011).
Crotty, S. T follicular helper cell biology: a decade of discovery and diseases. Immunity 50, 1132–1148 (2019).
Vinuesa, C. G., Linterman, M. A., Yu, D. & MacLennan, I. C. Follicular helper T cells. Annu. Rev. Immunol. 34, 335–368 (2016).
Webb, L. M. C. & Linterman, M. A. Signals that drive T follicular helper cell formation. Immunology 152, 185–194 (2017).
Wu, T. et al. TCF1 is required for the T follicular helper cell response to viral infection. Cell Rep. 12, 2099–2110 (2015).
Baumjohann, D., Okada, T. & Ansel, K. M. Cutting edge: distinct waves of BCL6 expression during T follicular helper cell development. J. Immunol. 187, 2089–2092 (2011).
Choi, Y. S. et al. ICOS receptor instructs T follicular helper cell versus effector cell differentiation via induction of the transcriptional repressor Bcl6. Immunity 34, 932–946 (2011).
DiToro, D. et al. Differential IL-2 expression defines developmental fates of follicular versus nonfollicular helper T cells. Science 361, eaao2933 (2018).
Tubo, N. J. et al. Single naive CD4+ T cells from a diverse repertoire produce different effector cell types during infection. Cell 153, 785–796 (2013).
Fazilleau, N., McHeyzer-Williams, L. J., Rosen, H. & McHeyzer-Williams, M. G. The function of follicular helper T cells is regulated by the strength of T cell antigen receptor binding. Nat. Immunol. 10, 375–384 (2009).
Krishnamoorthy, V. et al. The IRF4 gene regulatory module functions as a read–write integrator to dynamically coordinate T helper cell fate. Immunity 47, 481–497.e7 (2017).
Snook, J. M., Kim, C. & Williams, M. A. TCR signal strength controls the differentiation of CD4+ effector and memory T cells. Sci. Immunol. 3, eaas9103 (2018).
Keck, S. et al. Antigen affinity and antigen dose exert distinct influences on CD4 T-cell differentiation. Proc. Natl Acad. Sci. USA 111, 14852–14857 (2014).
Ploquin, M. J., Eksmond, U. & Kassiotis, G. B cells and TCR avidity determine distinct functions of CD4+ T cells in retroviral infection. J. Immunol. 187, 3321–3330 (2011).
Stefanova, I., Dorfman, J. R. & Germain, R. N. Self-recognition promotes the foreign antigen sensitivity of naive T lymphocytes. Nature 420, 429–434 (2002).
Persaud, S. P., Parker, C. R., Lo, W. L., Weber, K. S. & Allen, P. M. Intrinsic CD4+ T cell sensitivity and response to a pathogen are set and sustained by avidity for thymic and peripheral complexes of self peptide and MHC. Nat. Immunol. 15, 266–274 (2014).
van Oers, N. S. et al. Constitutive tyrosine phosphorylation of the T-cell receptor (TCR) ζ subunit: regulation of TCR-associated protein tyrosine kinase activity by TCR ζ. Mol. Cell. Biol. 13, 5771–5780 (1993).
Daley, S. R. et al. Rasgrp1 mutation increases naive T-cell CD44 expression and drives mTOR-dependent accumulation of Helios+ T cells and autoantibodies. Elife 2, e01020 (2013).
van Oers, N. S., Killeen, N. & Weiss, A. ZAP-70 is constitutively associated with tyrosine-phosphorylated TCR ζ in murine thymocytes and lymph node T cells. Immunity 1, 675–685 (1994).
Myers, D. R. et al. Tonic LAT–HDAC7 signals sustain Nur77 and Irf4 expression to tune naive CD4 T cells. Cell Rep. 19, 1558–1571 (2017).
Markegard, E. et al. Basal LAT–diacylglycerol–RasGRP1 signals in T cells maintain TCRα gene expression. PLoS ONE 6, e25540 (2011).
Zinzow-Kramer, W. M., Weiss, A. & Au-Yeung, B. B. Adaptation by naive CD4+ T cells to self-antigen-dependent TCR signaling induces functional heterogeneity and tolerance. Proc. Natl Acad. Sci. USA 116, 15160–15169 (2019).
Milam, A. A. V. et al. Tonic signaling inversely regulates the basal metabolism of CD4+ T cells. Immunohorizons 4, 485–497 (2020).
Azzam, H. S. et al. CD5 expression is developmentally regulated by T cell receptor (TCR) signals and TCR avidity. J. Exp. Med. 188, 2301–2311 (1998).
Zikherman, J., Parameswaran, R. & Weiss, A. Endogenous antigen tunes the responsiveness of naive B cells but not T cells. Nature 489, 160–164 (2012).
Moran, A. E. et al. T cell receptor signal strength in Treg and iNKT cell development demonstrated by a novel fluorescent reporter mouse. J. Exp. Med. 208, 1279–1289 (2011).
Bhandoola, A. et al. Peripheral expression of self-MHC-II influences the reactivity and self-tolerance of mature CD4+ T cells: evidence from a lymphopenic T cell model. Immunity 17, 425–436 (2002).
Viehmann Milam, A. A. et al. Tuning T cell signaling sensitivity alters the behavior of CD4+ T cells during an immune response. J. Immunol. 200, 3429–3437 (2018).
McHeyzer-Williams, L. J. & McHeyzer-Williams, M. G. Developmentally distinct TH cells control plasma cell production in vivo. Immunity 20, 231–242 (2004).
Weber, K. S. et al. Distinct CD4+ helper T cells involved in primary and secondary responses to infection. Proc. Natl Acad. Sci. USA 109, 9511–9516 (2012).
Pepper, M., Pagan, A. J., Igyarto, B. Z., Taylor, J. J. & Jenkins, M. K. Opposing signals from the Bcl6 transcription factor and the interleukin-2 receptor generate T helper 1 central and effector memory cells. Immunity 35, 583–595 (2011).
Yu, D. et al. The transcriptional repressor Bcl-6 directs T follicular helper cell lineage commitment. Immunity 31, 457–468 (2009).
Johnston, R. J. et al. Bcl6 and Blimp-1 are reciprocal and antagonistic regulators of T follicular helper cell differentiation. Science 325, 1006–1010 (2009).
Nurieva, R. I. et al. Bcl6 mediates the development of T follicular helper cells. Science 325, 1001–1005 (2009).
Zhu, J., Yamane, H. & Paul, W. E. Differentiation of effector CD4 T cell populations. Annu. Rev. Immunol. 28, 445–489 (2010).
Valderrama, C., Clark, A., Urano, F., Unanue, E. R. & Carrero, J. A. Listeria monocytogenes induces an interferon-enhanced activation of the integrated stress response that is detrimental for resolution of infection in mice. Eur. J. Immunol. 47, 830–840 (2017).
Good-Jacobson, K. L. & Shlomchik, M. J. Plasticity and heterogeneity in the generation of memory B cells and long-lived plasma cells: the influence of germinal center interactions and dynamics. J. Immunol. 185, 3117–3125 (2010).
Frey, A., Di Canzio, J. & Zurakowski, D. A statistically defined endpoint titer determination method for immunoassays. J. Immunol. Methods 221, 35–41 (1998).
Ahmed, N. N., Grimes, H. L., Bellacosa, A., Chan, T. O. & Tsichlis, P. N. Transduction of interleukin-2 antiapoptotic and proliferative signals via Akt protein kinase. Proc. Natl Acad. Sci. USA 94, 3627–3632 (1997).
Govern, C. C., Paczosa, M. K., Chakraborty, A. K. & Huseby, E. S. Fast on-rates allow short dwell time ligands to activate T cells. Proc. Natl Acad. Sci. USA 107, 8724–8729 (2010).
Mandl, J. N., Monteiro, J. P., Vrisekoop, N. & Germain, R. N. T cell-positive selection uses self-ligand binding strength to optimize repertoire recognition of foreign antigens. Immunity 38, 263–274 (2013).
Lo, W. L., Donermeyer, D. L. & Allen, P. M. A voltage-gated sodium channel is essential for the positive selection of CD4+ T cells. Nat. Immunol. 13, 880–887 (2012).
Tubo, N. J. et al. Most microbe-specific naive CD4+ T cells produce memory cells during infection. Science 351, 511–514 (2016).
Bocharov, G., Argilaguet, J. & Meyerhans, A.Understanding experimental LCMV infection of mice: the role of mathematical models. J. Immunol. Res. 2015, 739706 (2015).
Fung-Leung, W. P. et al. Antigen presentation and T cell development in H2–M-deficient mice. Science 271, 1278–1281 (1996).
Miyazaki, T. et al. Mice lacking H2–M complexes, enigmatic elements of the MHC class II peptide-loading pathway. Cell 84, 531–541 (1996).
Martin, W. D. et al. H2–M mutant mice are defective in the peptide loading of class II molecules, antigen presentation, and T cell repertoire selection. Cell 84, 543–550 (1996).
Brundage, R. A., Smith, G. A., Camilli, A., Theriot, J. A. & Portnoy, D. A. Expression and phosphorylation of the Listeria monocytogenes ActA protein in mammalian cells. Proc. Natl Acad. Sci. USA 90, 11890–11894 (1993).
Barber, D. L. et al. Restoring function in exhausted CD8 T cells during chronic viral infection. Nature 439, 682–687 (2006).
Acknowledgements
We thank D. Kreamalmeyer for mouse breeding and care, D. Brinja and E. Lantelme for assistance with sorting by flow cytometry, D. Oakley and M. Shih of the Washington University Center for Cellular Imaging for assistance with AxioScan use and image analysis, W. L. Lo for insights on the manuscript and M. White and the Genome Engineering and iPSC Center at Washington University for generation of the H2-DMaf/f mouse line. This work was supported by National Institutes of Health research grants AI138393 (to J.M.B.), AI130152-01A1 (to T.E.), AI139875 (to T.E.) and AI139540 (to P.M.A), the Leukemia and Lymphoma Society Scholar Award (to T.E.) and the Washington University Rheumatic Diseases Research Resource-Based Center, funded by NIH grant P30AR073752 (to P.M.A).
Author information
Authors and Affiliations
Contributions
J.M.B. designed and performed the experiments and wrote the manuscript. A.A.V.M., D.L.D., S.H., Y.X. and T.E. performed the experiments. P.M.A. designed the experiments and wrote the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Peer review information Zoltan Fehervari was the primary editor on this article and managed its editorial process and peer review in collaboration with the rest of the editorial team.
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Supplementary Information
Supplementary Figs. 1–4.
Rights and permissions
About this article
Cite this article
Bartleson, J.M., Viehmann Milam, A.A., Donermeyer, D.L. et al. Strength of tonic T cell receptor signaling instructs T follicular helper cell–fate decisions. Nat Immunol 21, 1384–1396 (2020). https://doi.org/10.1038/s41590-020-0781-7
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41590-020-0781-7
This article is cited by
-
Tuning charge density of chimeric antigen receptor optimizes tonic signaling and CAR-T cell fitness
Cell Research (2023)
-
TCR-pMHC: Envisioning the specialized dynamics of the target 5-component complex
Cellular & Molecular Immunology (2022)
-
A RORγt+ cell instructs gut microbiota-specific Treg cell differentiation
Nature (2022)
-
TCR-pMHC: may the force be of you?
Cellular & Molecular Immunology (2021)