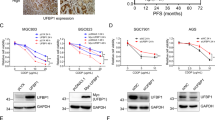
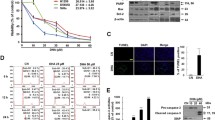
Abstract
Ablation of protein acyltransferase DHHC3 selectively enhanced the anti-cancer cell activities of several chemotherapeutic agents, but not kinase inhibitors. To understand why this occurs, we used comparative mass spectrometry-based palmitoyl-proteomic analysis of breast and prostate cancer cell lines, ± DHHC3 ablation, to obtain the first comprehensive lists of candidate protein substrates palmitoylated by DHHC3. Putative substrates included 22–28 antioxidant/redox-regulatory proteins, thus predicting that DHHC3 should have antioxidant functions. Consistent with this, DHHC3 ablation elevated oxidative stress. Furthermore, DHHC3 ablation, together with chemotherapeutic drug treatment, (a) elevated oxidative stress, with a greater than additive effect, and (b) enhanced the anti-growth effects of the chemotherapeutic agents. These results suggest that DHHC3 ablation enhances chemotherapeutic drug potency by disabling the antioxidant protections that contribute to drug resistance. Affirming this concept, DHHC3 ablation synergized with another anti-cancer drug, PARP inhibitor PJ-34, to decrease cell proliferation and increase oxidative stress. Hence, DHHC3 targeting can be a useful strategy for selectively enhancing potency of oxidative stress-inducing anti-cancer drugs. Also, comprehensive identification of DHHC3 substrates provides insight into other DHHC3 functions, relevant to in vivo tumor growth modulation.





Similar content being viewed by others
Availability of data and materials
Datasets generated and analyzed during the current study will be available as supplemental tables, and data with additional details will be available in the appropriate repository.
Abbreviations
- DHHC3:
-
Enzyme #3 in a family containing Asp–His–His–Cys motif
- NPC1:
-
NPC intracellular cholesterol transporter 1
- PalmPISC:
-
Palmitoyl protein identification and site characterization
- PARP:
-
Poly (ADP-ribose) polymerase
- PRDX4:
-
Peroxiredoxin 4
- SILAC:
-
Stable isotope labeling by amino acids in cell culture
- TMEM192:
-
Transmembrane protein 192
- TMX1 and TMX3:
-
Thioredoxin related transmembrane proteins 1 and 3
References
Mitchell DA, Vasudevan A, Linder ME, Deschenes RJ (2006) Protein palmitoylation by a family of DHHC protein S-acyltransferases. J Lipid Res 47:1118–1127
Yeste-Velasco M, Linder ME, Lu YJ (2015) Protein S-palmitoylation and cancer. Biochim Biophys Acta 1856:107–120
Ko PJ, Dixon SJ (2018) Protein palmitoylation and cancer. EMBO Rep 19:e46666
Sharma C, Wang HX, Li Q, Knoblich K, Reisenbichler ES, Richardson A, Hemler ME (2017) Protein acyltransferase DHHC3 regulates breast tumor growth, oxidative stress and senescence. Can Res 77:6880–6890
Yang W, Di Vizio D, Kirchner M, Steen H, Freeman MR (2010) Proteome scale characterization of human S-acylated proteins in lipid raft-enriched and non-raft membranes. Mol Cell Proteomics 9:54–70
Blagoev B, Kratchmarova I, Ong SE, Nielsen M, Foster LJ, Mann M (2003) A proteomics strategy to elucidate functional protein-protein interactions applied to EGF signaling. Nat Biotechnol 21:315–318
Cox J, Mann M (2008) MaxQuant enables high peptide identification rates, individualized p.p.b.-range mass accuracies and proteome-wide protein quantification. Nat Biotechnol 26:1367–1372
Cox J, Neuhauser N, Michalski A, Scheltema RA, Olsen JV, Mann M (2011) Andromeda: a peptide search engine integrated into the MaxQuant environment. J Proteome Res 10:1794–1805
Xu Q, Biener-Ramanujan E, Yang W, Ramanujan VK (2015) Targeting metabolic plasticity in breast cancer cells via mitochondrial complex I modulation. Breast Cancer Res Treat 150:43–56
Minciacchi VR, You S, Spinelli C, Morley S, Zandian M, Aspuria PJ, Cavallini L, Ciardiello C, Reis SM, Morello M, Kharmate G, Jang SC, Kim DK, Hosseini-Beheshti E, Tomlinson GE, Gleave M, Gho YS, Mathivanan S, Yang W, Freeman MR, Di VD (2015) Large oncosomes contain distinct protein cargo and represent a separate functional class of tumor-derived extracellular vesicles. Oncotarget 6:11327–11341
Burr ML, Sparbier CE, Chan YC, Williamson JC, Woods K, Beavis PA, Lam EYN, Henderson MA, Bell CC, Stolzenburg S, Gilan O, Bloor S, Noori T, Morgens DW, Bassik MC, Neeson PJ, Behren A, Darcy PK, Dawson SJ, Voskoboinik I, Trapani JA, Cebon J, Lehner PJ, Dawson MA (2017) CMTM6 maintains the expression of PD-L1 and regulates anti-tumour immunity. Nature 549:101–105
Fu R, Yanjanin NM, Bianconi S, Pavan WJ, Porter FD (2010) Oxidative stress in Niemann-Pick disease, type C. Mol Genet Metab 101:214–218
Chandiramani N, Wang X, Margeta M (2011) Molecular basis for vulnerability to mitochondrial and oxidative stress in a neuroendocrine CRI-G1 cell line. PLoS ONE 6:e14485
Liu Z, Lv YJ, Song YP, Li XH, Du YN, Wang CH, Hu LK (2012) Lysosomal membrane protein TMEM192 deficiency triggers crosstalk between autophagy and apoptosis in HepG2 hepatoma cells. Oncol Rep 28:985–991
Nystrom T, Yang J, Molin M (2012) Peroxiredoxins, gerontogenes linking aging to genome instability and cancer. Genes Dev 26:2001–2008
Hou D, Liu Z, Xu X, Liu Q, Zhang X, Kong B, Wei JJ, Gong Y, Shao C (2018) Increased oxidative stress mediates the antitumor effect of PARP inhibition in ovarian cancer. Redox Biol 17:99–111
Slade D (2019) Mitotic functions of poly(ADP-ribose) polymerases. Biochem Pharmacol 167:33–43
Kilpatrick CL, Murakami S, Feng M, Wu X, Lal R, Chen G, Du K, Luscher B (2016) Dissociation of Golgi-associated DHHC-type zinc finger protein (GODZ)- and Sertoli cell gene with a zinc finger domain-beta (SERZ-beta)-mediated palmitoylation by loss of function analyses in knock-out mice. J Biol Chem 291:27371–27386
Sharma C, Rabinovitz I, Hemler ME (2012) Palmitoylation by DHHC3 is critical for the function, expression and stability of integrin a6b4. Cell Mol Life Sci 69:2233–2244
Tsutsumi R, Fukata Y, Noritake J, Iwanaga T, Perez F, Fukata M (2009) Identification of G protein alpha subunit-palmitoylating enzyme. Mol Cell Biol 29:435–447
Keller CA, Yuan X, Panzanelli P, Martin ML, Alldred M, Sassoe-Pognetto M, Luscher B (2004) The gamma2 subunit of GABA(A) receptors is a substrate for palmitoylation by GODZ. J Neurosci 24:5881–5891
Wang J, Xie Y, Wolff DW, Abel PW, Tu Y (2010) DHHC protein-dependent palmitoylation protects regulator of G-protein signaling 4 from proteasome degradation. FEBS Lett 584:4570–4574
Lu D, Sun HQ, Wang H, Barylko B, Fukata Y, Fukata M, Albanesi JP, Yin HL (2012) Phosphatidylinositol 4-kinase IIalpha is palmitoylated by Golgi-localized palmitoyl transferases in cholesterol-dependent manner. J Biol Chem 287:21856–21865
Jennings BC, Linder ME (2012) DHHC protein S-acyltransferases use similar ping-pong kinetic mechanisms but display different acyl-CoA specificities. J Biol Chem 287:7236–7245
Linder ME, Deschenes RJ (2007) Palmitoylation: policing protein stability and traffic. Nat Rev Mol Cell Biol 8:74–84
Aicart-Ramos C, Valero RA, Rodriguez-Crespo I (2011) Protein palmitoylation and subcellular trafficking. Biochim Biophys Acta 1808:2981–2994
Handy DE, Loscalzo J (2012) Redox regulation of mitochondrial function. Antioxid Redox Signal 16:1323–1367
Gorrini C, Harris IS, Mak TW (2013) Modulation of oxidative stress as an anticancer strategy. Nat Rev Drug Discov 12:931–947
Trachootham D, Alexandre J, Huang P (2009) Targeting cancer cells by ROS-mediated mechanisms: a radical therapeutic approach? Nat Rev Drug Discov 8:579–591
Zhang X, Gibhardt CS, Will T, Stanisz H, Korbel C, Mitkovski M, Stejerean I, Cappello S, Pacheu-Grau D, Dudek J, Tahbaz N, Mina L, Simmen T, Laschke MW, Menger MD, Schon MP, Helms V, Niemeyer BA, Rehling P, Vultur A, Bogeski I (2019) Redox signals at the ER-mitochondria interface control melanoma progression. EMBO J 38:e100871
Raturi A, Gutierrez T, Ortiz-Sandoval C, Ruangkittisakul A, Herrera-Cruz MS, Rockley JP, Gesson K, Ourdev D, Lou PH, Lucchinetti E, Tahbaz N, Zaugg M, Baksh S, Ballanyi K, Simmen T (2016) TMX1 determines cancer cell metabolism as a thiol-based modulator of ER-mitochondria Ca2+ flux. J Cell Biol 214:433–444
Mezzadra R, Sun C, Jae LT, Gomez-Eerland R, de Vries, Wu W, Logtenberg MEW, Slagter M, Rozeman EA, Hofland I, Broeks A, Horlings HM, Wessels LFA, Blank CU, Xiao Y, Heck AJR, Borst J, Brummelkamp TR, Schumacher TNM (2017)Identification of CMTM6 and CMTM4 as PD-L1 protein regulators. Nature 549:106–110
Yao H, Lan J, Li C, Shi H, Brosseau JP, Wang H, Lu H, Fang C, Zhang Y, Liang L, Zhou X, Wang C, Xue Y, Cui Y, Xu J (2019) Inhibiting PD-L1 palmitoylation enhances T-cell immune responses against tumours. Nat Biomed Eng 3:306–317
Yang Y, Hsu JM, Sun L, Chan LC, Li CW, Hsu JL, Wei Y, Xia W, Hou J, Qiu Y, Hung MC (2019) Palmitoylation stabilizes PD-L1 to promote breast tumor growth. Cell Res 29:83–86
Landriscina M, Maddalena F, Laudiero G, Esposito F (2009) Adaptation to oxidative stress, chemoresistance, and cell survival. Antioxid Redox Signal 11:2701–2716
Funding
This study was supported by NIH grant CA42368 to M Hemler and C Sharma, DoD grant W81XWH-11-1-0112 to M Hemler, and DoD grant W81XWH-11-1-0113 to M Freeman. The funding body played no role in the design of the study, collection, analysis, and interpretation of data, or in writing the manuscript.
Author information
Authors and Affiliations
Contributions
WY and HS carried out mass spectrometry experiments, assisted by CS. CS carried out all other experiments. MF helped to direct the project. CS prepared figures and tables and wrote the manuscript, with assistance from MEH.
Corresponding authors
Ethics declarations
Conflict of interest
There are no financial or non-financial competing or conflicting interests to be declared.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Sharma, C., Yang, W., Steen, H. et al. Antioxidant functions of DHHC3 suppress anti-cancer drug activities. Cell. Mol. Life Sci. 78, 2341–2353 (2021). https://doi.org/10.1007/s00018-020-03635-3
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00018-020-03635-3