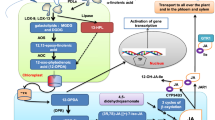
Key message
This study focused on the role of CLE1-CLE7 peptides as environmental mediators and indicated that root-induced CLE2 functions systemically in light-dependent carbohydrate metabolism in shoots.
Abstract
Plants sense environmental stimuli and convert them into cellular signals, which are transmitted to distinct cells and tissues to induce adequate responses. Plant hormones and small secretory peptides often function as environmental stress mediators. In this study, we investigated whether CLAVATA3/EMBRYO SURROUNDING REGION-RELATED proteins, CLE1–CLE7, which share closely related CLE domains, mediate environmental stimuli in Arabidopsis thaliana. Expression analysis of CLE1–CLE7 revealed that these genes respond to different environmental stimuli, such as nitrogen deprivation, nitrogen replenishment, cold, salt, dark, and sugar starvation, in a sophisticated manner. To further investigate the function of CLE2, we generated transgenic Arabidopsis lines expressing the β-glucuronidase gene under the control of the CLE2 promoter or expressing the CLE2 gene under the control of an estradiol-inducible promoter. We also generated cle2-1 and cle2-2 mutants using the CRISPR/Cas9 technology. In these transgenic lines, dark induced the expression of CLE2 in the root vasculature. Additionally, induction of CLE2 in roots induced the expression of various genes not only in roots but also in shoots, and genes related to light-dependent carbohydrate metabolism were particularly induced in shoots. In addition, cle2 mutant plants showed chlorosis when subjected to a shade treatment. These results suggest that root-induced CLE2 functions systemically in light-dependent carbohydrate metabolism in shoots.





Similar content being viewed by others
Data availability
The National Center for Biotechnology Information (NCBI) Gene Expression Omnibus (GEO) accession number for the microarray data reported in this paper is GSE149015.
Change history
02 April 2021
A Correction to this paper has been published: https://doi.org/10.1007/s11103-021-01144-w
References
Aoki Y, Okamura Y, Tadaka S, Kinoshita K, Obayashi T (2016) ATTED-II in 2016: a plant coexpression database towards lineage-specific coexpression. Plant Cell Physiol 57:e5
Araújo WL, Tohge T, Ishizaki K, Leaver CJ, Fernie AR (2011) Protein degradation–an alternative respiratory substrate for stressed plants. Trends Plant Sci 16:489–498
Araya T, Miyamoto M, Wibowo J, Suzuki A, Kojima S, Tsuchiya YN, Sawa S, Fukuda H, Wirén NV, Takahashi H (2014) CLE-CLAVATA1 peptide-receptor signaling module regulates the expansion of plant root systems in a nitrogen-dependent manner. Proc Natl Acad Sci USA 111:2029–2034
Baena-González E, Sheen J (2008) Convergent energy and stress signaling. Trends Plant Sci 13:474–482
Bidadi H, Matsuoka K, Sage-Ono K, Fukushima J, Pitaksaringkarn W, Asahina M, Yamaguchi S, Sawa S, Fukuda H, Matsubayashi Y, Ono M, Satoh S (2014) CLE6 expression recovers gibberellin deficiency to promote shoot growth in Arabidopsis. Plant J 78:241–252
Bonetta D, McCourt P (1998) Genetic analysis of ABA signal transduction pathways. Trends Plant Sci 3:231–235
Bray EA (1997) Plant responses to water deficit. Trends Plant Sci 2:48–53
Curtis MD, Grossniklaus U (2003) A gateway cloning vector set for high-throughput functional analysis of genes in planta. Plant Physiol 133:462–469
DiGennaro P, Grienenberger E, Dao TQ, Jun JH, Fletcher JC (2018) Peptide signaling molecules CLE5 and CLE6 affect Arabidopsis leaf shape downstream of leaf patterning transcription factors and auxin. Plant Direct 2:e00103
Endo S, Iwai Y, Fukuda H (2019) Cargo-dependent and cell wall-associated xylem transport in Arabidopsis. New Phytol 222:159–170
Endo S, Shinohara H, Matsubayashi Y, Fukuda H (2013) A novel pollen-pistil interaction conferring high-temperature tolerance during reproduction via CLE45 signaling. Curr Biol 23:1670–1676
Finkelstein RR, Gampala SSL, Rock CD (2002) Abscisic acid signaling in seeds and seedlings. Plant Cell 14:S15–S45
Fujiki Y, Yoshikawa Y, Sato T, Inada N, Ito M, Nishida I, Watanabe A (2001) Dark-inducible genes from Arabidopsis thaliana are associated with leaf senescence and repressed by sugars. Physiol Plant 111:345–352
Huang DW, Sherman BT, Zheng X, Yang J, Imamichi T, Stephens R, Lempicki RA (2009) Extracting biological meaning from large gene lists with DAVID. Curr Protoc Bioinformatics 27:11–13
Ichimura Y, Kirisako T, Takao T, Satomi Y, Shimonishi Y, Ishihara N, Mizushima N, Tanida I, Kominami E, Ohsumi M, Noda T, Ohsumi Y (2000) A ubiquitin-like system mediates protein lipidation. Nature 408:488–492
Ishida H, Yoshimoto K, Izumi M, Reisen D, Yano Y, Makino A, Ohsumi Y, Hanson MR and Mae T (2008) Mobilization of rubisco and stroma-localized fluorescent proteins of chloroplasts to the vacuole by an ATG gene-dependent autophagic process. Plant Physiol 148: 142–155
Ito Y, Nakanomyo I, Motose H, Iwamoto K, Sawa S, Dohmae N, Fukuda H (2006) Dodeca-CLE peptides as suppressors of plant stem cell differentiation. Science 313:842–845
Jun J, Fiume E, Roeder AH, Meng L, Sharma VK, Osmont KS, Baker C, Ha CM, Meyerowitz EM, Feldman LJ, Fletcher JC (2010) Comprehensive analysis of CLE polypeptide signaling gene expression and overexpression activity in Arabidopsis. Plant Physiol 154:1721–1736
Kinoshita A, Nakamura Y, Sasaki E, Kyozuka J, Fukuda H, Sawa S (2007) Gain-of-function phenotypes of chemically synthetic CLAVATA3/ESR-related (CLE) peptides in Arabidopsis thaliana and Oryza sativa. Plant Cell Physiol 48:1821–1825
Kondo Y, Hirakawa Y, Kieber JJ, Fukuda H (2011) CLE peptides can negatively regulate protoxylem vessel formation via cytokinin signaling. Plant Cell Physiol 52:37–48
Kucukoglu M, Nilsson O (2015) CLE peptide signaling in plants–the power of moving around. Physiol Plant 155:74–87
Matsubayashi Y (2011) Small post-translationally modified peptide signals in Arabidopsis. Arabidopsis Book 9:e0150
McDowell N, Pockman WT, Allen CD, Breshears DD, Cobb N, Kolb T, Plaut J, Sperry J, West A, Williams DG, Yepez EA (2008) Mechanisms of plant survival and mortality during drought: why do some plants survive while others succumb to drought? New Phytol 178:719–739
Nakatogawa H, Ichimura Y, Ohsumi Y (2007) Atg8, a ubiquitin-like protein required for autophagosome formation, mediates membrane tethering and hemifusion. Cell 130:165–178
Ni J, Clark SE (2006) Evidence for functional conservation, sufficiency, and proteolytic processing of the CLAVATA3 CLE domain. Plant Physiol 140:726–733
Obayashi T, Hayashi S, Saeki M, Ohta H, Kinoshita K (2009) ATTED-II provides coexpressed gene networks for Arabidopsis. Nucleic Acids Res 37:D987–D991
Oelkers K, Goffard N, Weiller GF, Gresshoff PM, Mathesius U, Frickey T (2008) Bioinformatic analysis of the CLE signaling peptide family. BMC Plant Biol 8:1471–2229
Ohashi-Ito K, Saegusa M, Iwamoto K, Oda Y, Katayama H, Kojima M, Sakakibara H, Fukuda H (2014) A bHLH complex activates vascular cell division via cytokinin action in root apical meristem. Curr Biol 24:2053–2058
Ohyama K, Shinohara H, Ogawa-Ohnishi M, Matsubayashi Y (2009) A glycopeptide regulating stem cell fate in Arabidopsis thaliana. Nat Chem Biol 5:578–580
Okamoto S, Ohnishi E, Sato S, Takahashi H, Nakazono M, Tabata S, Kawaguchi M (2009) Nod factor/nitrate-induced CLE genes that drive HAR1-mediated systemic regulation of nodulation. Plant Cell Physiol 50:67–77
Ono Y, Wada S, Izumi M, Makino A, Ishida H (2013) Evidence for contribution of autophagy to Rubisco degradation during leaf senescence in Arabidopsis thaliana. Plant Cell Environ 36:1147–1159
Polge C, Thomas M (2007) SNF1/AMPK/SnRK1 kinases, global regulators at the heart of energy control? Trends Plant Sci 12:20–28
Ramon M, Rolland F (2007) Plant development: introducing trehalose metabolism. Trends Plant Sci 12:185–188
Saito M, Kondo Y, Fukuda H (2018) BES1 and BZR1 redundantly promote phloem and xylem differentiation. Plant Cell Physiol 59:590–600
Scheible WR, Morcuende R, Czechowski T, Fritz C, Osuna D, Palacios-Rojas N, Schindelasch D, Thimm O, Udvardi MK, Stitt M (2004) Genome-wide reprogramming of primary and secondary metabolism, protein synthesis, cellular growth processes, and the regulatory infrastructure of Arabidopsis in response to nitrogen. Plant Physiol 136:2483–2499
Schwember AR, Schulze J, Del Pozo A, Cabeza RA (2019) Regulation of symbiotic nitrogen fixation in legume root nodules. Plants 8:333
Shinozaki K, Yamaguchi-Shinozaki K (1997) Gene expression and signal transduction in water-stress response. Plant Physiol 115:327–334
Shinozaki K, Yamaguchi-Shinozaki K (2000) Molecular responses to dehydration and low temperature: differences and cross-talk between two stress signaling pathways. Curr Opin Plant Biol 3:217–223
Strabala TJ, O'Donnell PJ, Smit AM, Ampomah-Dwamena C, Martin EJ, Netzler N, Nieuwenhuizen NJ, Quinn BD, Foote HC, Hudson KR (2006) Gain-of-function phenotypes of many CLAVATA3/ESR genes, including four new family members, correlate with tandem variations in the conserved CLAVATA3/ESR domain. Plant Physiol 140:1331–1344
Tabata R, Sumida K, Yoshii T, Ohyama K, Shinohara H, Matsubayashi Y (2014) Perception of root-derived peptides by shoot LRR-RKs mediates systemic N-demand signaling. Science 346:343–346
Takahashi F, Suzuki T, Osakabe Y, Betsuyaku S, Kondo Y, Dohmae N, Fukuda H, Yamaguchi-Shinozaki K, Shinozaki K (2018) A small peptide modulates stomatal control via abscisic acid in long-distance signalling. Nature 556:235–238
Verma V, Ravindran P, Kumar PP (2016) Plant hormone-mediated regulation of stress responses. BMC Plant Biol 16:86
Wada S, Ishida H, Izumi M, Yoshimoto K, Ohsumi Y, Mae T, Makino A (2009) Autophagy plays a role in chloroplast degradation during senescence in individually darkened leaves. Plant Physiol 149:885–893
Wang S, Blumwald E (2014) Stress-induced chloroplast degradation in Arabidopsis is regulated via a process independent of autophagy and senescence-associated vacuoles. Plant Cell 26:4875–4888
Yamaguchi YL, Ishida T, Yoshimura M, Imamura Y, Shimaoka C, Sawa S (2017) A collection of mutants for CLE-peptide-encoding genes in Arabidopsis generated by CRISPR/Cas9-mediated gene targeting. Plant Cell Physiol 58:1848–1856
Zhang H, Lin X, Han Z, Qu LJ, Chai J (2016) Crystal structure of PXY-TDIF complex reveals a conserved recognition mechanism among CLE peptide-receptor pairs. Cell Res 26:543–555
Zhang L, Shi X, Zhang Y, Wang J, Yang J, Ishida T, Jiang W, Han X, Kang J, Wang X, Pan L, Lv S, Cao B, Zhang Y, Wu J, Han H, Hu Z, Cui L, Sawa S, He J, Wang G (2019) CLE9 peptide-induced stomatal closure is mediated by abscisic acid, hydrogen peroxide, and nitric oxide in Arabidopsis thaliana. Plant Cell Environ 42:1033–1044
Acknowledgements
We thank Yukiko Sugisawa and Kuninori Iwamoto for technical support. We also thank Kyoko Ohashi-Ito, Yuki Kondo, Tomoyuki Furuya, Yumi Iwai and Alif Meem Nurani for discussion. This work was supported in part by Grants-in-Aid from the Ministry of Education, Culture, Sports, Science and Technology (15H05958) and from Japan Society for the Promotion of Science (16H06377) to HF.
Funding
This work was supported in part by Grant-in-Aid from the Ministry of Education, Culture, Sports, Science and Technology (15H05958) and from the Japan Society for the Promotion of Science (16H06377) to HF.
Author information
Authors and Affiliations
Contributions
DM, SE, SB, AS and HF designed the research; DM and SE performed the research; DM, SE and HF wrote the manuscript.
Corresponding authors
Ethics declarations
Conflicts of interest
The authors declare that they have no conflict of interest.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
The original version of this article was revised: the online Supplementary file1 (PDF 36 kb) was incomplete.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Ma, D., Endo, S., Betsuyaku, S. et al. CLE2 regulates light-dependent carbohydrate metabolism in Arabidopsis shoots. Plant Mol Biol 104, 561–574 (2020). https://doi.org/10.1007/s11103-020-01059-y
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11103-020-01059-y