Abstract
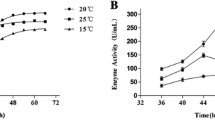
Thermal activity and stability are important characteristics for proteases applied in the detergent, pharmaceutical, food, and other green industries. With the intent to discover thermostable novel proteases, we constructed a fosmid metagenomic library from marine sediments in the East China Sea and isolated a clone endowed with high proteolytic activity from this library. Sequence analysis of the positive subclones allowed the identification of a coding region of 1254 bp related to protease activity. The unrooted phylogenetic tree and alignment results revealed that the sequence might be derived from Anaerolineaceae bacterium and encodes a new member of the peptidase S8A subfamily with the typical catalytic triad Asp119/His150/Ser325. The fusion protein, named pF1AL2, was expressed in Escherichia coli and showed a molecular weight of 35 kDa. pF1AL2 was active in the pH range of 5.0–11.0 with an optimal pH at 10.0 and had high stability under alkaline conditions, retaining more than 95% of its activity after 24 h at pH 11.0. The optimal temperature of pF1AL2 was 80 °C, and it retained nearly 80% of its activity after 6 h at 70 °C, showing great thermal activity and stability. In addition, the enzyme had great salt tolerance (the residual activity when kept in 3 M NaCl was 40%). Its thermal activity and stability, along with its halotolerance and pH-tolerance, indicate the high potential value of pF1AL2 in industrial applications. The exploitation of pF1AL2 could lay the foundation for the development and utilization of proteases with special features from marine resources by a metagenomic strategy.
Key points
• A novel protease, pF1AL2, from marine sediments, was screened out by a metagenomic strategy.
• The protease pF1AL2 analyzed in silico, cloned, and characterized.
• pF1AL2 had an optimal temperature of 80 °C and retained nearly 80% of activity after 6 h at 70 °C.
• pF1AL2 had great tolerance for high-temperature and acid, alkaline, and high salt environments.





Similar content being viewed by others
References
Apolinar-Hernández MM, Peña-Ramírez YJ, Pérez-Rueda E, Canto-Canché BB, Santos-Briones DC, O'Connor-Sánchez A (2016) Identification and in silico characterization of two novel genes encoding peptidases S8 found by functional screening in a metagenomic library of Yucatán underground water. Gene 593(1):154–161
Barzkar N, Homaei A, Hemmati R, Seema P (2018) Thermostable marine microbial proteases for industrial applications: scopes and risks. Extremophiles 22:335–346
Berlemont R, Pipers D, Delsaute M, Angiono F, Feller G, Galleni M, Power P (2011) Exploring the Antarctic soil metagenome as a source of novel cold-adapted enzymes and genetic mobile elements. Rev Argent Microbiol 43(2):94–103
Biver S, Portetelle D, Vandenbol M (2013) Characterization of a new oxidant-stable serine protease isolated by functional metagenomics. Springerplus 2(1):410
Bouzas TdM, Barros-Velázquez J, Villa TG (2006) Industrial applications of hyperthermophilic enzymes: a review. Protein Pept Lett 13(7):645–651
Chatterjee J, Giri S, Maity S, Sinha A, Ranjan A, Rajshekhar GS (2015) Production and characterization of thermostable alkaline protease of Bacillus subtilis (ATCC 6633) from optimized solid-state fermentation. Biotechnol Appl Biochem 62(5):709–718
Chu XM, He HZ, Guo CQ, Sun BL (2008) Identification of two novel esterases from a marine metagenomic library derived from South China Sea. Appl Microbiol Biotechnol 80:615–625
Devi SG, Fathima AA, Sanitha M, Iyappan S, Curtis WR, Ramya M (2016) Expression and characterization of alkaline protease from the metagenomic library of tannery activated sludge. J Biosci Bioeng 122(6):694–700
Felczykowska A, Bloch SK, Nejman-Faleńczyk B, Barańska S (2012) Metagenomic approach in the investigation of new bioactive compounds in the marine environment. Acta Biochim Pol 59(4):501–505
Gohel SD, Singh SP (2018) Thermodynamics of a Ca2+ dependent, highly thermostable and detergent compatible purified alkaline serine protease from Nocardiopsis xinjiangensis strain OM-6. Int J Biol Macromol 113:565–574
Gong BL, Mao RQ, Xiao Y, Jia ML, Zhong XL, Liu Y, Xu PL, Li G (2017) Improvement of enzyme activity and soluble expression of an alkaline protease isolated from oil-polluted mud flat metagenome by random mutagenesis. Enzym Microb Technol 106:97–105
Guo YJ, Tu T, Zheng J, Ren YX, Wang YR, Bai YG, Su XY, Wang Y, Yao B, Huang HQ, Luo HY (2020) A novel thermostable aspartic protease from Talaromyces leycettanus and its specific autocatalytic activation through an intermediate transition state. Appl Microbiol Biotechnol 104:4915–4926. https://doi.org/10.1007/s00253-020-10569-0
Jiang C, Zhang L, Li F, Meng C, Zeng R, Deng J, Shen P, Ou Q, Wu B (2017) Characterization of a metagenome-derived protease from contaminated agricultural soil microorganisms and its random mutagenesis. Folia Microbiol (Praha) 62(6):499–508
Kennedy J, Marchesi JR, Dobson AD (2008) Marine metagenomics: strategies for the discovery of novel enzymes with biotechnological applications from marine environments. Microb Cell Factories 7:27
Lee D-G, Jeon JH, Jang MK, Kim NY, Lee JH, Lee J-H, Kim S-J, Kim G-D, Lee S-H (2007) Screening and characterization of a novel fibrinolytic metalloprotease from a metagenomic library. Biotechnol Lett 29(3):465–472
Martin M, Vandenbol M (2016) The hunt for original microbial enzymes: An initiatory review on the construction and functional screening of (meta)genomic libraries. Biotechnol Agron Soc Environ 20(4):523–532
Morris LS, Marchesi JR (2015) Current functional metagenomic approaches only expand the existing protease sequence space, but does not presently add any novelty to it. Curr Microbiol 70(1):19–26
Neurath H (1989) Proteolytic processing and physiological regulation. Trends Biochem Sci 14(7):268–271
Neveu J, Regeard C, DuBow MS (2011) Isolation and characterization of two serine proteases from metagenomic libraries of the Gobi and Death Valley deserts. Appl Microbiol Biotechnol 91(3):635–644
Niehaus F, Gabor E, Wieland S, Siegert P, Maurer KH, Eck J (2011) Enzymes for the laundry industries: tapping the vast metagenomic pool of alkaline proteases. Microb Biotechnol 4(6):767–776
Pessoa TBA, Rezende RP, Marques EDLS, Pirovani CP, Santos TFD, Gonçalves ACDS, Romano CC, Dotivo NC, Freitas ACO, Salay LC, Dias JCT (2017) Metagenomic alkaline protease from mangrove sediment. J Basic Microbiol 57(11):962–973
Petersen TN, Brunak S, Heijne GV, Nielsen H (2011) SignalP 4.0: discriminating signal peptides from transmembrane regions. Nat Methods 8(10):785–786
Potumarthi R, Ch S, Jetty A (2007) Alkaline protease production by submerged fermentation in stirred tank reactor using Bacillus licheniformis NCIM-2042: effect of aeration and agitation regimes. Biochem Eng J 34:185–192
Purohit MK, Singh SP (2013) A metagenomic alkaline protease from saline habitat: cloning, over-expression and functional attributes. Int J Biol Macromol 53(2):138–143
Pushpam PL, Rajesh T, Gunasekaran P (2011) Identification and characterization of alkaline serine protease from goat skin surface metagenome. AMB Express 1(1):3
Rawlings ND, Barrett AJ, Finn R (2016) Twenty years of the MEROPS database of proteolytic enzymes, their substrates and inhibitors. Nucleic Acids Res 44(Database issue):D343–D350
Ribitsch D, Karl W, Birner-Gruenberger R, Gruber K, Eiteljoerg I, Remler P, Wieland S, Siegert P, Maurer KH, Schwab H (2010) C-terminal truncation of a metagenome-derived detergent protease for effective expression in E. coli. J Biotechnol 150(3):408–416
Schloss PD, Handelsman J (2003) Biotechnological prospects from metagenomics. Curr Opin Biotechnol 14(3):303–310
Siezen RJ, Leunissen JAM (1997) Subtilases: the superfamily of subtilisin-like serine proteases. Protein Sci 6(3):501–523
Singh R, Chopra C, Gupta VK, Akhlaq B, Verma V, Rasool S (2015) Purification and characterization of CHpro1, a thermotolerant, alkali-stable and oxidation-resisting protease of Chumathang hotspring. Sci Bull (Beijing) 60(14):1252–1260
Su C, Gong JS, Zhang RX, Tao LY, Dou WF, Zhang DD, Li H, Lu ZM, Xu ZH, Shi JS (2017) A novel alkaline surfactant-stable keratinase with superior feather-degrading potential based on library screening strategy. Int J Biol Macromol 95:404–411
Tamura K, Stecher G, Peterson D, Filipski A, Kumar S (2013) MEGA6: molecular evolutionary genetics analysis version 6.0. Mol Biol Evol 30(12):2725–2729
Tavano OL, Berenguer-Murcia A, Secundo F, Fernandez-Lafuente R (2018) Biotechnological applications of proteases in food technology. Compr Rev Food Sci Food Saf 00:1–25
Ueda K, Lipkind GM, Zhou A, Zhu X, Kuznetsov A, Philipson L, Gardner P, Zhang C, Steiner DF (2003) Mutational analysis of predicted interactions between the catalytic and P domains of prohormone convertase 3 (PC3/PC1). Proc Natl Acad Sci U S A 100(10):5622–5627
Wang HL, Hart DJ, An YF (2019) Functional metagenomic technologies for the discovery of novel enzymes for biomass degradation and biofuel production. Bioenerg Res 12:457–470
Whitman WB, Coleman DC, Wiebe WJ (1998) Prokaryotes: the unseen majority. Proc Natl Acad Sci U S A 95(12):6578–6583
Zhang Y, Zhao J, Zeng R (2011) Expression and characterization of a novel mesophilic protease from metagenomic library derived from Antarctic coastal sediment. Extremophiles 15(1):23–29
Funding
This work was supported by the China Agriculture Research System (CARS-48), the Beihai 13th 5-year marine economic innovation and development demonstration project (Bhsfs011), the Applied Basic Research Program of Qingdao (16-5-1-18-jch), and the Fundamental Research Funds for the Central Universities (201564018).
Author information
Authors and Affiliations
Contributions
Xiangzhao Mao conceived and designed research. Jianan Sun and Ping Li conducted experiments and wrote manuscript. Zhen Liu and Wencan Huang contributed new reagents and analytical tools. All authors read and approved the manuscript.
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Ethical statement
This article does not contain any studies with human participants or animals performed by any of the authors.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
ESM 1
(PDF 547 kb)
Rights and permissions
About this article
Cite this article
Sun, J., Li, P., Liu, Z. et al. A novel thermostable serine protease from a metagenomic library derived from marine sediments in the East China Sea. Appl Microbiol Biotechnol 104, 9229–9238 (2020). https://doi.org/10.1007/s00253-020-10879-3
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00253-020-10879-3