Abstract
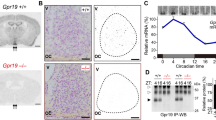
The mammalian circadian system consists of a major circadian pacemaker located in the suprachiasmatic nucleus (SCN) of the hypothalamus and peripheral clocks in the body, including brain structures. The SCN depends on glutamatergic neurotransmission for transmitting signals from the retina, and it exhibits spontaneous 24-h rhythmicity in neural activity. The aim of this work was to evaluate the degree and circadian rhythmicity of AMPA receptor GluA2 subunit R/G editing and alternative flip/flop splicing in the SCN and other brain structures in Wistar rats. Our data show that the circadian rhythmicity in the SCN’s GluA2 mRNA level was highest at dawn, while the circadian rhythm in R/G editing peaked at CT10 and the rhythmic flip varied with the acrophase at the late subjective night. The circadian rhythmicity was confirmed for R/G editing and splicing in the CA3 hippocampal area, and rhythmic variation of the flip isoform was also measured in the olfactory bulbs and cerebellum. The correlations between the R/G editing and alternative flip/flop splicing revealed a structure-dependent direction. In the hippocampus, the edited (G)-form level was positively correlated with the flip variant abundance, in accord with published data; by contrast, in the SCN, the flip variant was in associated more with the unedited (R) form. The edited (G) form and flop isoform also predominated in the retina and cerebellum.




Similar content being viewed by others
References
Hastings MH, Maywood ES, Brancaccio M (2018) Generation of circadian rhythms in the suprachiasmatic nucleus. Nat Rev Neurosci 19(8):453–469. https://doi.org/10.1038/s41583-018-0026-z
Rath MF, Rohde K, Møller M (2012) Circadian oscillations of molecular clock components in the cerebellar cortex of the rat. Chronobiol Int 29(10):1289–1299. https://doi.org/10.3109/07420528.2012.728660
Bering T, Carstensen MB, Wörtwein G, Weikop P, Rath MF (2018) The circadian oscillator of the cerebral cortex: molecular, biochemical and behavioral effects of deleting the Arntl clock gene in cortical neurons. Cereb Cortex 28(2):644–657. https://doi.org/10.1093/cercor/bhw406
Jilg A, Lesny S, Peruzki N, Schwegler H, Selbach O, Dehghani F, Stehle JH (2010) Temporal dynamics of mouse hippocampal clock gene expression support memory processing. Hippocampus 20(3):377–388. https://doi.org/10.1002/hipo.20637
Besharse JC, McMahon DG (2016) The retina and other light-sensitive ocular clocks. J Biol Rhythm 31(3):223–243. https://doi.org/10.1177/0748730416642657
Moriya T, Ikeda M, Teshima K, Hara R, Kuriyama K, Yoshioka T, Allen CN, Shibata S (2003) Facilitation of alpha-amino-3-hydroxy-5-methylisoxazole-4-propionate receptor transmission in the suprachiasmatic nucleus by aniracetam enhances photic responses of the biological clock in rodents. J Neurochem 85(4):978–987. https://doi.org/10.1046/j.1471-4159.2003.01758.x
Mizoro Y, Yamaguchi Y, Kitazawa R, Yamada H, Matsuo M, Fustin JM, Doi M, Okamura H (2010) Activation of AMPA receptors in the suprachiasmatic nucleus phase-shifts the mouse circadian clock in vivo and in vitro. PLoS One 5(6):e10951. https://doi.org/10.1371/journal.pone.0010951
Haak LL, Albers HE, Mintz EM (2006) Modulation of photic response by the metabotropic glutamate receptor agonist t-ACPD. Brain Res Bull 71(1–3):97–100. https://doi.org/10.1016/j.brainresbull.2006.08.006
Colwell CS, Menaker M (1992) NMDA as well as non-NMDA receptor antagonists can prevent the phase-shifting effects of light on the circadian system of the golden hamster. J Biol Rhythm 7:125–136. https://doi.org/10.1177/074873049200700204
Abe H, Rusak B, Robertson HA (1992) NMDA and non-NMDA receptor antagonists inhibit photic induction of Fos protein in the hamster suprachiasmatic nucleus. Brain Res Bull 28:831–835. https://doi.org/10.1007/BF00239919
Michel S, Itri J, Colwell CS (2002) Excitatory mechanisms in the suprachiasmatic nucleus: the role of AMPA/KA glutamate receptors. J Neurophysiol 88(2):817–828. https://doi.org/10.1152/jn.2002.88.2.817
Martin LJ, Blackstone CD, Levey AI, Huganir RL, Price DL (1993) AMPA glutamate receptor subunits are differentially distributed in rat brain. Neuroscience 53(2):327–358. https://doi.org/10.1016/0306-4522(93)90199-p
Eyigor O, Centers A, Jennes L (2001) Distribution of ionotropic glutamate receptor subunit mRNAs in the rat hypothalamus. J Comp Neurol 434(1):101–124. https://doi.org/10.1002/cne.1167
Pellegrini-Giampietro DE, Gorter JA, Bennett MV, Zukin RS (1997) The GluR2 (GluR-B) hypothesis: Ca(2+)-permeable AMPA receptors in neurological disorders. Trends Neurosci 20(10):464–470. https://doi.org/10.1016/s0166-2236(97)01100-4
Higuchi M, Maas S, Single FN, Hartner J, Rozov A, Burnashev N, Feldmeyer D, Sprengel R et al (2000) Point mutation in an AMPA receptor gene rescues lethality in mice deficient in the RNA-editing enzyme ADAR2. Nature 406(6791):78–81. https://doi.org/10.1038/35017558
Twomey EC, Yelshanskaya MV, Grassucci RA, Frank J, Sobolevsky AI (2017) Channel opening and gating mechanism in AMPA-subtype glutamate receptors. Nature 549(7670):60–65. https://doi.org/10.1038/nature23479
Krampfl K, Schlesinger F, Zörner A, Kappler M, Dengler R, Bufler J (2002) Control of kinetic properties of GluR2 flop AMPA-type channels: impact of R/G nuclear editing. Eur J Neurosci 15(1):51–62. https://doi.org/10.1046/j.0953-816x.2001.01841.x
Lomeli H, Mosbacher J, Melcher T, Hoger T, Geiger JR, Kuner T, Monyer H, Higuchi M et al (1994) Control of kinetic properties of AMPA receptor channels by nuclear RNA editing. Science 266:1709–1713. https://doi.org/10.1126/science.7992055
Greger IH, Akamine P, Khatri L, Ziff EB (2006) Developmentally regulated, combinatorial RNA processing modulates AMPA receptor biogenesis. Neuron 51:85–97. https://doi.org/10.1016/j.neuron.2006.05.020
Sommer B, Keinanen K, Verdoorn TA, Wisden W, Burnashev N, Herb A, Köhler M, Takagi T et al (1990) Flip and flop: a cell-specific functional switch in glutamate-operated channels of the CNS. Science. 249:1580–1585. https://doi.org/10.1126/science.1699275
Schoft VK, Schopoff S, Jantsch MF (2007) Regulation of glutamate receptor B pre-mRNA splicing by RNA editing. Nucleic Acids Res 35(11):3723–3732. https://doi.org/10.1093/nar/gkm314
Penn AC, Greger IH (2009) Sculpting AMPA receptor formation and function by alternative RNA processing. RNA Biol 6(5):517–521. https://doi.org/10.4161/rna.6.5.9552
Penn AC, Balik A, Wozny C, Cais O, Greger IH (2012) Activity-mediated AMPA receptor remodeling, driven by alternative splicing in the ligand-binding domain. Neuron 76(3):503–510. https://doi.org/10.1016/j.neuron.2012.08.010
Balik A, Penn AC, Nemoda Z, Greger IH (2013) Activity-regulated RNA editing in select neuronal subfields in hippocampus. Nucleic Acids Res 41(2):1124–1134. https://doi.org/10.1093/nar/gks1045
Sanjana NE, Levanon EY, Hueske EA, Ambrose JM, Li JB (2012) Activity-dependent A-to-I RNA editing in rat cortical neurons. Genetics. 192(1):281–287. https://doi.org/10.1534/genetics.112.141200
Bonini D, Filippini A, La Via L, Fiorentini C, Fumagalli F, Colombi M, Barbon A (2015) Chronic glutamate treatment selectively modulates AMPA RNA editing and ADAR expression and activity in primary cortical neurons. RNA Biol 12(1):43–53. https://doi.org/10.1080/15476286.2015.1008365
Wright A, Vissel B (2012) The essential role of AMPA receptor GluR2 subunit RNA editing in the normal and diseased brain. Front Mol Neurosci 5:34. https://doi.org/10.3389/fnmol.2012.00034
Colwell CS (2011) Linking neural activity and molecular oscillations in the SCN. Nat Rev Neurosci 12(10):553–569. https://doi.org/10.1038/nrn3086
Granados-Fuentes D, Saxena MT, Prolo LM, Aton SJ, Herzog ED (2004) Olfactory bulb neurons express functional, entrainable circadian rhythms. Eur J Neurosci 19(4):898–906. https://doi.org/10.1111/j.0953-816x.2004.03117.x
Huang H, Tan BZ, Shen Y, Tao J, Jiang F, Sung YY, Ng CK, Raida M et al (2012) RNA editing of the IQ domain in Ca(v)1.3 channels modulates their Ca2+-dependent inactivation. Neuron 73(2):304–316. https://doi.org/10.1016/j.neuron.2011.11.022
Terajima H, Yoshitane H, Yoshikawa T, Shigeyoshi Y, Fukada Y (2018) A-to-I RNA editing enzyme ADAR2 regulates light-induced circadian phase-shift. Sci Rep 8(1):14848. https://doi.org/10.1038/s41598-018-33114-6
Pačesová D, Novotný J, Bendová Z (2016) The effect of chronic morphine or methadone exposure and withdrawal on clock gene expression in the rat suprachiasmatic nucleus and AA-NAT activity in the pineal gland. Physiol Res 65(3):517–525. https://doi.org/10.33549/physiolres.933183
Ge B, Gurd S, Gaudin T, Dore C, Lepage P, Harmsen E, Hudson TJ, Pastinen T (2005) Survey of allelic expression using EST mining. Genome Res 15:1584–1591. https://doi.org/10.1101/gr.4023805
Hahnová K, Pačesová D, Volfová B, Červená K, Kašparová D, Žurmanová J, Bendová Z (2016) Circadian Dexras1 in rats: Development, location and responsiveness to light. Chronobiol Int 33(2):141–150. https://doi.org/10.3109/07420528.2015.1120741
Chambille I (1999) Circadian rhythm of AMPA receptor GluR2/3 subunit-immunoreactivity in the suprachiasmatic nuclei of Syrian hamster and effect of a light-dark cycle. Brain Res 833(1):27–38. https://doi.org/10.1016/s0006-8993(99)01460-2
Lundkvist GB, Kristensson K, Hill RH (2002) The suprachiasmatic nucleus exhibits diurnal variations in spontaneous excitatory postsynaptic activity. J Biol Rhythm 17(1):40–51. https://doi.org/10.1177/074873002129002320
Michel S, Marek R, Vanderleest HT, Vansteensel MJ, Schwartz WJ, Colwell CS, Meijer JH (2013) Mechanism of bilateral communication in the suprachiasmatic nucleus. Eur J Neurosci 37(6):964–971. https://doi.org/10.1111/ejn.12109
Grosskreutz J, Zoerner A, Schlesinger F, Krampfl K, Dengler R, Bufler J (2003) Kinetic properties of human AMPA-type glutamate receptors expressed in HEK293 cells. Eur J Neurosci 17:1173–1178. https://doi.org/10.1046/j.1460-9568.2003.02531.x
Wen W, Lin CY, Niu L (2017) R/G editing in GluA2Rflop modulates the functional difference between GluA1 flip and flop variants in GluA1/2R heteromeric channels. Sci Rep 7(1):13654. https://doi.org/10.1038/s41598-017-13233-2
Terajima H, Yoshitane H, Ozaki H, Suzuki Y, Shimba S, Kuroda S, Iwasaki W, Fukada Y (2017) ADARB1 catalyzes circadian A-to-I editing and regulates RNA rhythm. Nat Genet 49(1):146–151. https://doi.org/10.1038/ng.3731
Green DJ, Gillette R (1982) Circadian rhythm of firing rate recorded from single cells in the rat suprachiasmatic brain slice. Brain Res 245:198–200. https://doi.org/10.1016/0006-8993(82)90361-4
Groos G, Hendriks J (1982) Circadian rhythms in electrical discharge of rat suprachiasmatic neurons recorded in vitro. Neurosci Lett 34:283–288. https://doi.org/10.1016/0304-3940(82)90189-6
Bos NP, Mirmiran M (1990) Circadian rhythms in spontaneous neuronal discharges of the cultured suprachiasmatic nucleus. Brain Res 511:158–162. https://doi.org/10.1016/0006-8993(90)90235-4
Jones JR, Tackenberg MC, McMahon DG (2015) Manipulating circadian clock neuron firing rate resets molecular circadian rhythms and behavior. Nat Neurosci 18(3):373–375. https://doi.org/10.1038/nn.3937
Munn RG, Bilkey DK (2012) The firing rate of hippocampal CA1 place cells is modulated with a circadian period. Hippocampus 22(6):1325–1337. https://doi.org/10.1002/hipo.20969
Meijer JH, Schwartz WJ (2003) In search of the pathways for light-induced pacemaker resetting in the suprachiasmatic nucleus. J Biol Rhythm 18(3):235–249. https://doi.org/10.1177/0748730403018003006
LeSauter J, Silver R, Cloues R, Witkovsky P (2011) Light exposure induces short- and long-term changes in the excitability of retinorecipient neurons in suprachiasmatic nucleus. J Neurophysiol 106(2):576–588. https://doi.org/10.1152/jn.00060.2011
Zaidan H, Ramaswami G, Golumbic YN, Sher N, Malik A, Barak M, Galiani D, Dekel N et al (2018) A-to-I RNA editing in the rat brain is age-dependent, region-specific and sensitive to environmental stress across generations. BMC Genomics 19(1):28. https://doi.org/10.1186/s12864-017-4409-8
Dick A, Khermesh K, Paul E, Stamp F, Levano EY, Chen A (2019) Adenosine-to-inosine RNA editing within corticolimbic brain regions is regulated in response to chronic social defeat stress in mice. Front Psychiatry 10:277. https://doi.org/10.3389/fpsyt.2019.00277
Bonini D, Mora C, Tornese P, Sala N, Filippini A, La Via L, Milanese M, Calza S et al (2016) Acute footshock stress induces time-dependent modifications of AMPA/NMDA protein expression and AMPA phosphorylation. Neural Plast 7267865:1–10. https://doi.org/10.1155/2016/7267865
Kubota-Sakashita M, Iwamoto K, Bundo M, Kato T (2014) A role of ADAR2 and RNA editing of glutamate receptors in mood disorders and schizophrenia. Mol Brain 7:5. https://doi.org/10.1186/1756-6606-7-5
Barbon A, Vallini I, La Via L, Marchina E, Barlati S (2003) Glutamate receptor RNA editing: a molecular analysis of GluR2, GluR5 and GluR6 in human brain tissues and in NT2 cells following in vitro neural differentiation. Brain Res Mol Brain Res 117(2):168–178. https://doi.org/10.1016/s0169-328x(03)00317-6
Park YH, Broyles HV, He S, McGrady NR, Li L, Yorio T (2016) Involvement of AMPA receptor and its Flip and Flop isoforms in retinal ganglion cell death following oxygen/glucose deprivation. Invest Ophthalmol Vis Sci 57(2):508–526. https://doi.org/10.1167/iovs.15-18481
Lundin E, Wu C, Widmark A, Behm M, Hjerling-Leffler J, Daniel C, Öhman M, Nilsson M (2020) Spatiotemporal mapping of RNA editing in the developing mouse brain using in situ sequencing reveals regional and cell-type-specific regulation. BMC Biol 18(1):6. https://doi.org/10.1186/s12915-019-0736-3
Ravindranathan A, Donevan SD, Sugden SG, Greig A, Rao MS, Parks TN (2000) Contrasting molecular composition and channel properties of AMPA receptors on chick auditory and brainstem motor neurons. J Physiol 523(Pt 3):667–684. https://doi.org/10.1111/j.1469-7793.2000.00667.x
Jakobs T, Ben Y, Masland RH (2007) Expression of mRNA for glutamate receptor subunits distinguishes the major classes of retinal neurons, but is less specific for individual cell types. Mol Vis 13:933–948. http://www.molvis.org/molvis/v13/a99/
Corthell JT, Fadool DA, Trombley PQ (2012) Connexin and AMPA receptor expression changes over time in the rat olfactory bulb. Neuroscience 222:38–48. https://doi.org/10.1016/j.neuroscience.2012.06.070
Bratt E, Ohman M (2003) Coordination of editing and splicing of glutamate receptor pre-mRNA. RNA 9(3):309–318. https://doi.org/10.1261/rna.2750803
Penn AC, Balik A, Greger IH (2013) Steric antisense inhibition of AMPA receptor Q/R editing reveals tight coupling to intronic editing sites and splicing. Nucleic Acids Res 41(2):1113–1123. https://doi.org/10.1093/nar/gks1044
Patton AP, Hastings MH (2018) The suprachiasmatic nucleus. Curr Biol 28(15):R816–R822. https://doi.org/10.1016/j.cub.2018.06.052
Greger IH, Khatri L, Ziff EB (2002) RNA editing at arg607 controls AMPA receptor exit from the endoplasmic reticulum. Neuron 34(5):759–772. https://doi.org/10.1016/s0896-6273(02)00693-1
Funding
This work was supported by the Czech Science Foundation (grant no. 19-17037S); the Charles University Grant Agency (grant no. 1030217); the European Regional Development Fund-Projects “PharmaBrain” No. CZ.CZ.02.1.01/0.0/0.0/16_025/0007444; Research Project of the AS CR RVO: 67985823; the Ministry of Education, Youth and Sports of the Czech Republic: LQ1604 National Sustainability Program II (Project BIOCEV-FAR); and the “BIOCEV” project (CZ.1.05/1.1.00/02.0109).
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Conflict of Interest
The authors declare that they have no conflicts of interest.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Míková, H., Kuchtiak, V., Svobodová, I. et al. Circadian Regulation of GluA2 mRNA Processing in the Rat Suprachiasmatic Nucleus and Other Brain Structures. Mol Neurobiol 58, 439–449 (2021). https://doi.org/10.1007/s12035-020-02141-8
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12035-020-02141-8