Abstract
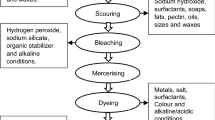
Textile industry consumes a large proportion of available water and releases huge amounts of toxic azo dye effluents, leading to an inevitable situation of acute environmental pollution that has been a significant threat to mankind. Decolorization or detoxification of harmful azo dyes has become a global priority to overcome the disastrous consequences and salvage the ecosystem. Biodegradation of textile azo dyes by endophytes stands to be a lucrative and viable alternative over conventional physico-chemical methods, owing to their eco-friendliness, cost-competitive and non-toxic nature. Especially, plant endophytic microbes exhibit promising biodegradation potential which has wired up the effective removal of textile azo dyes, attributing to their ability to produce dye degrading enzymes, laccases, peroxidases and azoreductases. Although both bacterial and fungal endophytes have been tried for azo dye degradation, endophytic fungi find broader application over bacteria. Despite of the advancements made in microbe-mediated biodegradation, there is still a need to fill the gap in lab to in situ translation of biodegradation research. This review concisely accentuates the xenobiotics of textile azo dyes and microbial mechanisms of biodegradation of textile azo dyes, positing plant endophytic community, especially bacterial and fungal endophytes as the potential dye degraders, highlighting currently reported dye degrading endophytic species.




Similar content being viewed by others
References
Desore A, Narula SA (2018) An overview on corporate response towards sustainability issues in textile industry. Environ Dev Sustain 20(4):1439–1459
Bhatia S (2017) Pollution control in textile industry. CRC Press, Boca Raton
Baban A, Yediler A, Lienert D, Kemerdere N, Kettrup A (2003) Ozonation of high strength segregated effluents from a woolen textile dyeing and finishing plant. Dyes Pigm 58(2):93–98
Bafana A, Devi SS, Chakrabarti T (2011) Azo dyes: past, present and the future. Environ Rev 19:350–371
Hossain MS, Das SC, Islam JM, Al Mamun MA, Khan MA (2018) Reuse of textile mill ETP sludge in environmental friendly bricks—effect of gamma radiation. Radiat Phys Chem 151:77–83
Rehman K, Shahzad T, Sahar A, Hussain S, Mahmood F, Siddique MH, Siddique MA, Rashid MI (2018) Effect of Reactive Black 5 azo dye on soil processes related to C and N cycling. PeerJ 6:e4802
Lellis B, Fávaro-Polonio CZ, Pamphile JA, Polonio JC (2019) Effects of textile dyes on health and the environment and bioremediation potential of living organisms. Biotechnol Res Innov 3:275–290
Rawat D, Mishra V, Sharma RS (2016) Detoxification of azo dyes in the context of environmental processes. Chemosphere 155:591–605
Imran M, Crowley DE, Khalid A, Hussain S, Mumtaz MW, Arshad M (2015) Microbial biotechnology for decolorization of textile wastewaters. Rev Environ Sci Bio/Technol 14(1):73–92
Gogate PR, Pandit AB (2004) A review of imperative technologies for wastewater treatment II: hybrid methods. Adv Environ Res 8(3–4):553–597
Sarkar S, Banerjee A, Halder U, Biswas R, Bandopadhyay R (2017) Degradation of synthetic azo dyes of textile industry: a sustainable approach using microbial enzymes. Water Conserv Sci Eng 2(4):121–131
Sudha M, Saranya A, Selvakumar G, Sivakumar N (2014) Microbial degradation of azo dyes: a review. Int J Curr Microbiol Appl Sci 3(2):670–690
Mishra S, Maiti A (2019) Applicability of enzymes produced from different biotic species for biodegradation of textile dyes. Clean Technol Environ Policy 21(4):763–781
Corrêa RCG, Iark D, de Sousa Idelfonso A, Uber TM, Bracht A, Peralta RM (2018) Endophytes as pollutant-degrading agents: current trends and perspectives. In Endophytes and secondary metabolites. Springer, Cham, pp 1–22
Sreedharan V, Rao KVB (2019) Biodegradation of textile azo dyes. In: Nanoscience and biotechnology for environmental applications. Springer, Cham, pp 115–139
Garg SK, Tripathi M (2017) Microbial strategies for discoloration and detoxification of azo dyes from textile effluents. Res J Microbiol 12:1–19
Sim CSF, Chen SH, Ting ASY (2019) Endophytes: emerging tools for the bioremediation of pollutants. In: Emerging and eco-friendly approaches for waste management. Springer, Singapore, pp 189–217
Mansour HB, Houas I, Montassar F, Ghedira K, Barillier D, Mosrati R, Chekir-Ghedira L (2012) Alteration of in vitro and acute in vivo toxicity of textile dyeing wastewater after chemical and biological remediation. Environ Sci Pollut Res 19(7):2634–2643
Sghaier I, Guembri M, Chouchane H (2019) Recent advances in textile wastewater treatment using microbial consortia. J Text Eng Fashion Technol 5(3):134–146
Barathi S, Arulselvi PI (2015) Decolorization, degradation, and toxicological analysis of textile dye effluent by using novel techniques—review. Int J Sci Res Manag 3(2):2118–2136
Chequer FMD, Angeli JPF, Ferraz ERA, Tsuboy MS, Marcarini JC, Mantovani MS, de Oliveira DP (2009) The azo dyes Disperse Red 1 and Disperse Orange 1 increase the micronuclei frequencies in human lymphocytes and in HepG2 cells. Mutat Res Genet Toxicol Environ Mutagenesis 676(1–2):83–86
Sivarajasekar N, Baskar R (2014) Adsorption of basic red 9 on activated waste Gossypium hirsutum seeds: process modeling, analysis and optimization using statistical design. J Ind Eng Chem 20(5):2699–2709
Piątkowska M, Jedziniak P, Olejnik M, Żmudzki J, Posyniak A (2018) Absence of evidence or evidence of absence? A transfer and depletion study of Sudan I in eggs. Food Chem 239:598–602
Christie RM (2007) Environmental aspects of textile dyeing. Elsevier, Amsterdam
Gopinath KP, Murugesan S, Abraham J, Muthukumar K (2009) Bacillus sp. mutant for improved biodegradation of Congo red: random mutagenesis approach. Bioresour Technol 100(24):6295–6300
Gottilieb A, Shaw C, Smith A, Wheatley A, Forsythe S (2003) The toxicity of textile reactive azo dyes after hydrolysis and decolourization. J Biotechnol 101(1):49–56
Chequer FMD, Lizier TM, de Felício R, Zanoni MVB, Debonsi HM, Lopes NP, de Oliveira DP (2015) The azo dye Disperse Red 13 and its oxidation and reduction products showed mutagenic potential. Toxicol In Vitro 29(7):1906–1915
Akansha K, Chakraborty D, Sachan SG (2019) Decolorization and degradation of methyl orange by Bacillus stratosphericus SCA1007. Biocatal Agric Biotechnol 18:101044
Sandesh K, Kumar G, Chidananda B, Ujwal P (2019) Optimization of Direct Blue-14 dye degradation by Bacillus fermus (Kx898362) an alkaliphilic plant endophyte and assessment of degraded metabolite toxicity. J Hazard Mater 364:742–751
Little L, Lamb III J (1973) Acute toxicity of 46 selected dyes to the Fathead Minnow, Pimephales promelas. In: Dyes and the environment-reports on selected dyes and their effects, vol 1. American Dye Manufacturers Institute, p 130
Mansour HB, Ayed-Ajmi Y, Mosrati R, Corroler D, Ghedira K, Barillier D, Chekir-Ghedira L (2010) Acid violet 7 and its biodegradation products induce chromosome aberrations, lipid peroxidation, and cholinesterase inhibition in mouse bone marrow. Environ Sci Pollut Res 17(7):1371–1378
Guo G, Tian F, Zhang C, Liu T, Yang F, Hu Z, Liu C, Wang S, Ding K (2019) Performance of a newly enriched bacterial consortium for degrading and detoxifying azo dyes. Water Sci Technol 79(11):2036–2045
Govindwar SP, Kurade MB, Tamboli DP, Kabra AN, Kim PJ, Waghmode TR (2014) Decolorization and degradation of xenobiotic azo dye Reactive Yellow-84A and textile effluent by Galactomyces geotrichum. Chemosphere 109:234–238
Bankole PO, Adekunle AA, Obidi OF, Chandanshive VV, Govindwar SP (2018) Biodegradation and detoxification of Scarlet RR dye by a newly isolated filamentous fungus, Peyronellaea prosopidis. Sustain Environ Res 28(5):214–222
Rodriguez RJ, Henson J, Van Volkenburgh E, Hoy M, Wright L, Beckwith F, Kim Y-O, Redman RS (2008) Stress tolerance in plants via habitat-adapted symbiosis. ISME J 2(4):404
Taghavi S, Barac T, Greenberg B, Borremans B, Vangronsveld J, van der Lelie D (2005) Horizontal gene transfer to endogenous endophytic bacteria from poplar improves phytoremediation of toluene. Appl Environ Microbiol 71(12):8500–8505
Arunkumara P, Soundarapandianb N (2017) Bioremediation of color and COD from dye effluent using indigenous and exogenous microbes. Desalin Water Treat 59:243–247
Glick BR, Stearns JC (2011) Making phytoremediation work better: maximizing a plant’s growth potential in the midst of adversity. Int J Phytoremediation 13(sup1):4–16
Shehzadi M, Fatima K, Imran A, Mirza M, Khan Q, Afzal M (2016) Ecology of bacterial endophytes associated with wetland plants growing in textile effluent for pollutant-degradation and plant growth-promotion potentials. Plant Biosyst 150(6):1261–1270
Kabra AN, Khandare RV, Govindwar SP (2013) Development of a bioreactor for remediation of textile effluent and dye mixture: a plant–bacterial synergistic strategy. Water Res 47(3):1035–1048
Patil SM, Chandanshive VV, Rane NR, Khandare RV, Watharkar AD, Govindwar SP (2016) Bioreactor with Ipomoea hederifolia adventitious roots and its endophyte Cladosporium cladosporioides for textile dye degradation. Environ Res 146:340–349
Wuhrmann K, Mechsner K, Kappeler T (1980) Investigation on rate—determining factors in the microbial reduction of azo dyes. Eur J Appl Microbiol Biotechnol 9(4):325–338
Dave SR, Patel TL, Tipre DR (2015) Bacterial degradation of azo dye containing wastes. In: Microbial degradation of synthetic dyes in wastewaters. Springer, Cham, pp 57–83
Imran M, Negm F, Hussain S, Ashraf M, Ashraf M, Ahmad Z, Arshad M, Crowley DE (2016) Characterization and purification of membrane‐bound azoreductase from azo dye degrading Shewanella sp. strain IFN4. Clean: Soil, Air, Water 44(11):1523–1530
Haghshenas H, Kay M, Dehghanian F, Tavakol H (2016) Molecular dynamics study of biodegradation of azo dyes via their interactions with AzrC azoreductase. J Biomol Struct Dyn 34(3):453–462
Shang N, Ding M, Dai M, Si H, Li S, Zhao G (2019) Biodegradation of malachite green by an endophytic bacterium Klebsiella aerogenes S27 involving a novel oxidoreductase. Appl Microbiol Biotechnol 103(5):2141–2153
Khandare R, Kabra A, Awate A, Govindwar SP (2013) Synergistic degradation of diazo dye Direct Red 5B by Portulaca grandiflora and Pseudomonas putida. Int J Environ Sci Technol 10(5):1039–1050
Shi X, Liu Q, Ma J, Liao H, Xiong X, Zhang K, Wang T, Liu X, Xu T, Yuan S (2015) An acid-stable bacterial laccase identified from the endophyte Pantoea ananatis Sd-1 genome exhibiting lignin degradation and dye decolorization abilities. Biotechnol Lett 37(11):2279–2288
Riva V, Mapelli F, Syranidou E, Crotti E, Choukrallah R, Kalogerakis N, Borin S (2019) Root bacteria recruited by Phragmites australis in constructed wetlands have the potential to enhance azo-dye phytodepuration. Microorganisms 7(10):384
Gayathri S, Saravanan D, Radhakrishnan M, Balagurunathan R, Kathiresan K (2010) Bioprospecting potential of fast growing endophytic bacteria from leaves of mangrove and salt-marsh plant species. Indian J Biotechnol 9(4):397–402
Pundir RK, Rana S, Kaur A, Kashyap N, Jain P (2014) Bioprospecting potential of endophytic bacteria isolated from indigenous plants of Ambala (Haryana, India). Int J Pharm Sci Res 5(6):2309
Masarbo RS, Niranjana S, Monisha T, Nayak AS, Karegoudar T (2019) Efficient decolorization and detoxification of sulphonated azo dye Ponceau 4R by using single and mixed bacterial consortia. Biocatal Biotransform 37(5):367–376
Guo G, Li X, Tian F, Liu T, Yang F, Ding K, Liu C, Chen J, Wang C (2020) Azo dye decolorization by a halotolerant consortium under microaerophilic conditions. Chemosphere 244:125510
Khalid A, Mahmood S (2015) The biodegradation of azo dyes by actinobacteria. In: Microbial degradation of synthetic dyes in wastewaters. Springer, Cham, pp 297–314
Lu L, Zeng G, Fan C, Ren X, Wang C, Zhao Q, Zhang J, Chen M, Chen A, Jiang M (2013) Characterization of a laccase-like multicopper oxidase from newly isolated Streptomyces sp. C1 in agricultural waste compost and enzymatic decolorization of azo dyes. Biochem Eng J 72:70–76
Moya R, Hernández M, García-Martín AB, Ball AS, Arias ME (2010) Contributions to a better comprehension of redox-mediated decolouration and detoxification of azo dyes by a laccase produced by Streptomyces cyaneus CECT 3335. Bioresour Technol 101(7):2224–2229
Shehzadi M, Afzal M, Khan MU, Islam E, Mobin A, Anwar S, Khan QM (2014) Enhanced degradation of textile effluent in constructed wetland system using Typha domingensis and textile effluent-degrading endophytic bacteria. Water Res 58:152–159
Shilpa S, Shikha R (2015) Biodegradation of dye reactive black-5 by a novel bacterial endophyte. Int Res J Environ Sci 4(4):44–53
Shahid M, Mahmood F, Hussain S, Shahzad T, Haider MZ, Noman M, Mushtaq A, Fatima Q, Ahmed T, Mustafa G (2018) Enzymatic detoxification of azo dyes by a multifarious Bacillus sp. strain MR-1/2-bearing plant growth-promoting characteristics. 3 Biotech 8(10):425
Libra JA, Borchert M, Banit S (2003) Competition strategies for the decolorization of a textile-reactive dye with the white-rot fungi Trametes versicolor under non-sterile conditions. Biotechnol Bioeng 82(6):736–744
Deng Z, Cao L (2017) Fungal endophytes and their interactions with plants in phytoremediation: a review. Chemosphere 168:1100–1106
Humnabadkar R, Saratale G, Govindwar SP (2008) Decolorization of purple 2R by Aspergillus ochraceus (NCIM-1146). Asian J Microbiol Biotechnol Environ Exp Sci 10(3):693–697
Bergsten-Torralba LR, Nishikawa MM, Baptista DF, Magalhães DdP, Silva Md (2009) Decolorization of different textile dyes by Penicillium simplicissimum and toxicity evaluation after fungal treatment. Braz J Microbiol 40(4):808–817
Singh S, Pakshirajan K (2010) Enzyme activities and decolourization of single and mixed azo dyes by the white-rot fungus Phanerochaete chrysosporium. Int Biodeterior Biodegrad 64(2):146–150
Kaushik P, Malik A (2015) Mycoremediation of synthetic dyes: an insight into the mechanism, process optimization and reactor design. In: Microbial degradation of synthetic dyes in wastewaters. Springer, Cham, pp 1–25
Ting ASY, Lee MVJ, Chow YY, Cheong SL (2016) Novel exploration of endophytic Diaporthe sp. for the biosorption and biodegradation of triphenylmethane dyes. Water Air Soil Pollut 227(4):109
Ngieng NS, Zulkharnain A, Roslan HA, Husaini A (2013) Decolourisation of synthetic dyes by endophytic fungal flora isolated from senduduk plant (Melastoma malabathricum). ISRN Biotechnol 2013:18
Tochhawng L, Mishra VK, Passari AK, Singh BP (2019) Endophytic fungi: role in dye decolorization. In: Advances in endophytic fungal research. Springer, Cham, pp 1–15
Gahlout M, Gupte S, Gupte A (2013) Optimization of culture condition for enhanced decolorization and degradation of azo dye reactive violet 1 with concomitant production of ligninolytic enzymes by Ganoderma cupreum AG-1. 3 Biotech 3(2):143–152
Sidhu A, Agrawal S, Sable V, Patil S, Gaikwad V (2014) Isolation of Colletotrichum gloeosporioides gr., a novel endophytic laccase producing fungus from the leaves of a medicinal plant, Piper betle. Int J Sci Eng Res 5:1087–1096
Muthezhilan R, Vinoth S, Gopi K, Jaffar Hussain A (2014) Dye degrading potential of immobilized laccase from endophytic fungi of coastal sand dune plants. Int J ChemTech Res 6(9):4154–4160
Verma SK, Kumar A, Lal M, Debnath M (2015) Biodegradation of synthetic dye by endophytic fungal isolate in Calotropis procera root. Int J Appl Sci Biotechnol 3(3):373–380
Agrawal S (2017) Decolorization of textile dye by laccase from newly isolated endophytic fungus Daldinia sp. Kavaka 48:33–41
Bulla LMC, Polonio JC, de Brito Portela-Castro AL, Kava V, Azevedo JL, Pamphile JA (2017) Activity of the endophytic fungi Phlebia sp. and Paecilomyces formosus in decolourisation and the reduction of reactive dyes’ cytotoxicity in fish erythrocytes. Environ Monit Assess 189(2):88
Sun J, Guo N, Niu L-L, Wang Q-F, Zang Y-P, Zu Y-G, Fu Y-J (2017) Production of laccase by a new Myrothecium verrucaria MD-R-16 isolated from Pigeon Pea [Cajanus cajan (L.) Millsp.] and its application on dye decolorization. Molecules 22(4):673
Marzall-Pereira M, Savi DC, Bruscato EC, Niebisch CH, Paba J, Aluízio R, Ferreira-Maba LS, Galli-Terasawa LV, Glienke C, Kava V (2019) Neopestalotiopsis species presenting wide dye destaining activity: Report of a mycelium-associated laccase. Microbiol Res 228:126299
Jin R, Yang H, Zhang A, Wang J, Liu G (2009) Bioaugmentation on decolorization of CI Direct Blue 71 by using genetically engineered strain Escherichia coli JM109 (pGEX-AZR). J Hazard Mater 163(2–3):1123–1128
Liu G-f, Zhou J-t, Qu Y-y, Ma X (2007) Decolorization of sulfonated azo dyes with two photosynthetic bacterial strains and a genetically engineered Escherichia coli strain. World J Microbiol Biotechnol 23(7):931–937
Rathod J, Dhebar S, Archana G (2017) Efficient approach to enhance whole cell azo dye decolorization by heterologous overexpression of Enterococcus sp. L2 azoreductase (azoA) and Mycobacterium vaccae formate dehydrogenase (fdh) in different bacterial systems. Int Biodeterior Biodegrad 124:91–100
Liang Y, Hou J, Liu Y, Luo Y, Tang J, Cheng JJ, Daroch M (2018) Textile dye decolorizing Synechococcus PCC7942 engineered with CotA laccase. Front Bioeng Biotechnol 6:95
Pandey G, Paul D, Jain RK (2005) Conceptualizing “suicidal genetically engineered microorganisms” for bioremediation applications. Biochem Biophys Res Commun 327(3):637–639
Gadd GM (2009) Biosorption: critical review of scientific rationale, environmental importance and significance for pollution treatment. J Chem Technol Biotechnol 84(1):13–28
Sharma S, Saxena R, Gaur G (2014) Study of removal techniques for azo dyes by biosorption: a review. J Appl Chem 7(10):06–21
Ahalya N, Ramachandra T, Kanamadi R (2003) Biosorption of heavy metals. Res J Chem Environ 7(4):71–79
Abdi O, Kazemi M (2015) A review study of biosorption of heavy metals and comparison between different biosorbents. J Mater Environ Sci 6(5):1386–1399
Takey M, Shaikh T, Mane N, Majumder D (2014) Bioremidiation of xenobiotics: use of dead fungal biomass as biosorbent. Int J Res Eng Technol 3(1):565–570
Saraf S, Vaidya VK (2015) Comparative study of biosorption of textile dyes using fungal biosorbents. Int J Curr Microbiol App Sci 2:357–365
Vijayaraghavan K, Yun Y-S (2008) Polysulfone-immobilized Corynebacterium glutamicum: a biosorbent for Reactive black 5 from aqueous solution in an up-flow packed column. Chem Eng J 145(1):44–49
Vijayaraghavan K, Yun Y-S (2008) Bacterial biosorbents and biosorption. Biotechnol Adv 26(3):266–291
Aksu Z, Dönmez G (2005) Combined effects of molasses sucrose and reactive dye on the growth and dye bioaccumulation properties of Candida tropicalis. Process Biochem 40(7):2443–2454
Aksu Z (2003) Reactive dye bioaccumulation by Saccharomyces cerevisiae. Process Biochem 38(10):1437–1444
Husain Q (2010) Peroxidase mediated decolorization and remediation of wastewater containing industrial dyes: a review. Rev Environ Sci Bio/Technol 9(2):117–140
Robinson T, McMullan G, Marchant R, Nigam P (2001) Remediation of dyes in textile effluent: a critical review on current treatment technologies with a proposed alternative. Bioresour Technol 77(3):247–255
Saratale RG, Saratale GD, Chang J-S, Govindwar SP (2011) Bacterial decolorization and degradation of azo dyes: a review. J Taiwan Inst Chem Eng 42(1):138–157
Telke AA, Kadam AA, Govindwar SP (2015) Bacterial enzymes and their role in decolorization of azo dyes. In: Microbial degradation of synthetic dyes in wastewaters. Springer, Cham, pp 149–168
Keck A, Klein J, Kudlich M, Stolz A, Knackmuss H-J, Mattes R (1997) Reduction of azo dyes by redox mediators originating in the naphthalenesulfonic acid degradation pathway of Sphingomonas sp. strain BN6. Appl Environ Microbiol 63(9):3684–3690
Chacko JT, Subramaniam K (2011) Enzymatic degradation of azo dyes—a review. Int J Environ Sci 1(6):1250
Pandey A, Singh P, Iyengar L (2007) Bacterial decolorization and degradation of azo dyes. Int Biodeterior Biodegrad 59(2):73–84
Lima DR, Baeta BE, Silva GAd, Silva SQ, Aquino SF (2014) Use of multivariate experimental designs for optimizing the reductive degradation of an azo dye in the presence of redox mediators. Quím Nova 37(5):827–832
Yang J, Yang X, Lin Y, Ng TB, Lin J, Ye X (2015) Laccase-catalyzed decolorization of malachite green: performance optimization and degradation mechanism. PLoS ONE 10(5):e0127714
Zheng F, Cui B-K, Wu X-J, Meng G, Liu H-X, Si J (2016) Immobilization of laccase onto chitosan beads to enhance its capability to degrade synthetic dyes. Int Biodeterior Biodegrad 110:69–78
Chen H (2006) Recent advances in azo dye degrading enzyme research. Curr Protein Pept Sci 7(2):101–111
Pereira L, Coelho AV, Viegas CA, dos Santos MMC, Robalo MP, Martins LO (2009) Enzymatic biotransformation of the azo dye Sudan Orange G with bacterial CotA-laccase. J Biotechnol 139(1):68–77
Durao P, Bento I, Fernandes AT, Melo EP, Lindley PF, Martins LO (2006) Perturbations of the T1 copper site in the CotA laccase from Bacillus subtilis: structural, biochemical, enzymatic and stability studies. J Biol Inorg Chem 11(4):514
Mate DM, Alcalde M (2015) Laccase engineering: from rational design to directed evolution. Biotechnol Adv 33(1):25–40
Abedin RM (2008) Decolorization and biodegradation of crystal violet and malachite green by Fusarium solani (Martius) Saccardo. A comparative study on biosorption of dyes by the dead fungal biomass. Am Euras J Bot 12:17–31
Zille A, Gornacka B, Rehorek A, Cavaco-Paulo A (2005) Degradation of azo dyes by Trametes villosa laccase over long periods of oxidative conditions. Appl Environ Microbiol 71(11):6711–6718
Roriz MS, Osma JF, Teixeira JA, Couto SR (2009) Application of response surface methodological approach to optimise Reactive Black 5 decolouration by crude laccase from Trametes pubescens. J Hazard Mater 169(1–3):691–696
Mendes S, Robalo MP, Martins LO (2015) Bacterial enzymes and multi-enzymatic systems for cleaning-up dyes from the environment. In: Microbial degradation of synthetic dyes in wastewaters. Springer, Cham, pp 27–55
Durán N, Rosa MA, D’Annibale A, Gianfreda L (2002) Applications of laccases and tyrosinases (phenoloxidases) immobilized on different supports: a review. Enzym Microb Technol 31(7):907–931
Gomare SS, Jadhav JP, Govindwar SP (2008) Degradation of sulfonated azo dyes by the purified lignin peroxidase from Brevibacillus laterosporus MTCC 2298. Biotechnol Bioprocess Eng 13(2):136–143
McMullan G, Meehan C, Conneely A, Kirby N, Robinson T, Nigam P, Banat I, Marchant R, Smyth W (2001) Microbial decolourisation and degradation of textile dyes. Appl Microbiol Biotechnol 56(1–2):81–87
Acknowledgements
This research was financially supported by the Ministry of Small and Medium-sized Enterprises (SMEs) and Startups (MSS), Korea, under the “Regional Specialized Industry Development Program (R&D, S2931078)” supervised by the Korea Institute for Advancement of Technology (KIAT). This work was supported by “Human Resources Program in Energy Technology” of the Korea Institute of Energy Technology Evaluation and Planning (KETEP), granted financial resource from the Ministry of Trade, Industry & Energy, Republic of Korea (No. 20204010600100).
Author information
Authors and Affiliations
Contributions
BSG has the idea for the article, performed the literature search and data analysis, BSG and GK have drafted, designed the figures, and critically revised the entire work, HLC has also performed the certain literature search and JHK has supervised the entire work.
Corresponding authors
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Goud, B.S., Cha, H.L., Koyyada, G. et al. Augmented Biodegradation of Textile Azo Dye Effluents by Plant Endophytes: A Sustainable, Eco-Friendly Alternative. Curr Microbiol 77, 3240–3255 (2020). https://doi.org/10.1007/s00284-020-02202-0
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00284-020-02202-0