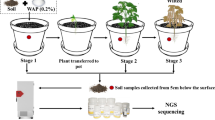
Abstract
Bacteria are the most abundant soil microbes and are sensitive to environmental change, especially soil carbon (C) and nitrogen (N) dynamics. The bacterial diversity of rhizosphere and bulk soils associated with desert plants is not well understood. In this study, we measured the properties of rhizosphere and bulk soils at different depths (0–20, 20–40, 40–60, and 60–80 cm), the diversity of bacterial communities (16S rDNA amplicon sequencing), and their relationships with Anabasis aphylla in the southern margin of the Gurbantunggut Desert, Junggar Basin, China. A total of 11,420 operational taxonomic units (OTUs) were obtained from 40 soil samples, belonging to 641 genera, 269 families, 137 orders, 61 classes, and 44 phyla. There were significant differences in electrical conductivity (EC), available nitrogen (AN), available phosphorus (AP), available potassium (AK), and bacterial diversity. The dominant bacterial communities of the rhizosphere and bulk soils at the phylum level were Actinobacteria, Proteobacteria, and Bacteroidetes. At the genus level, the dominant communities of the rhizosphere and bulk soils were Halomonas and Glycomyces, respectively. At different soil depths, the abundances of bacteria in the soil were 10.2% (0–20 cm) > 8.4% (20–40 cm) > 8.3% (60–80 cm) > 6.2% (40–60 cm). Our results indicate that bacteria in the phyla Actinobacteria and Proteobacteria, as well as the genus Halomonas, are key to the drought and salt tolerance of A. aphylla.






Similar content being viewed by others
References
Haichar FEZ, Santaella C, Heulin T, Achouak W (2014) Root exudates mediated interactions belowground. Soil Biol Biochem 77:69–80
Loon LCV (2007) Plant responses to plant-growth promoting rhizbacteria. Eur J Plant Pathol 119(3):243–254
Philippot L, Raaijmakers JM, Lemanceau P, Van der Putten WH, (2013) Going back to the roots: the microbial ecology of the rhizosphere. Nat Rev Microbiol, 11(11): 789-799
Prashar P, Kapoor N, Sachdeva S (2014) Rhizosphere: its structure, bacterial diversity and significance. Rev Environ Sci Biotechnol 13(1):63–77
Delgado-Baquerizo M, Reich PB, Khachane AN, Campbell CD, Thomas N, Freitag TE, Abu Al-Soud W, Sorensen S, Bardgett RD, Singh BK (2017) It is elemental: soil nutrient stoichiometry drives bacterial diversity. Environ Microbiol 19(3):1176–1188
Raaijmakers JM, Vlami M, de Souza JT (2002) Antibiotic production by bacterial biocontrol agents. Anton Leeuw Int J G 81(1–4):537–547
Lipson DA, Blair M, Barron-Gafford G, Grieve K, Murthy R (2006) Relationships between microbial community structure and soil processes under elevated atmospheric carbon dioxide. Microb Ecol 51(3):302–314
Maul JE, Buyer JS, Lehman RM, Culman S, Teasdale JR (2014) Microbial community structure and abundance in the rhizosphere and bulk soil of a tomato cropping system that includes cover crops. Appl Soil Ecol 77:42–50
Kang YJ, Cheng J, Mei LJ, Yin SX (2010) Screening and identification of plant rhizosphere-promoting rhizobacteria. Acta Microbiol Sin 50(7):853–861 (in Chinese)
Chu GM, Wang M, Zhang SX (2014) Spatial patterns and associations of dominant woody species in desert-oasis ecotone of South Junggar Basin NW China. J Plant Interact 9(1):738–744
Hassler M (2019) World plants: synonymic checklists of the vascular plants of the world (version Nov 2018).
Bray SR, Kitajima K, Mack MC (2012) Temporal dynamics of microbial communities on decomposing leaf litter of 10 plant species in relation to decomposition rate. Soil Biol Biochem 49:30–37
Feng Y, Zhang ML, Zhuo L, Chen MZ (2011) Micromorphology of Anabasis from China with reference to their ecological significance. Acta Bot Boreal Occident Sin 31(3):462–467 (in Chinese)
Peng MW, Chang YL, Chu GM, Wang M (2019) Low-temperature tolerance and transcriptome analyses during seed germination of Anabasis aphylla. J Plant Interact 14(1):254–264
Li XM, Zuo YL, Xue ZK, Zhang LL, He XL (2018) Characteristics of rhizosphere soil microbial community structure of different desert plants. Acta Ecol Sin 38(8):2855–2863 (in Chinese)
Yang Y, Liu BR (2015) Distribution of soil nutrient and microbial biomass in rhizosphere versus non-rhizosphere area of different plant species in desertified steppe. Acta Ecol Sin 35(22):7562–7570 (in Chinese)
Yang ZA, Zhu QA, Zhan W, Xu YY, Zhu EX, Gao YH, Li SQ, Zheng QY, Zhu D, He YX, Peng CH, Huai C (2018) The linkage between vegetation and soil nutrients and their variation under different grazing intensities in an alpine meadow on the eastern Qinghai-Tibetan Plateau. Ecol Eng 110:128–136
Chaudhary DR, Gautam RK, Yousuf B, Mishra A, Jha B (2015) Nutrients, microbial community structure and functional gene abundance of rhizosphere and bulk soils of halophytes. Appl Soil Ecol 91:16–26
Zhang C, Liu GB, Xue S, Zhang CS (2012) Rhizosphere soil microbial properties on abandoned croplands in the Loess Plateau, China during vegetation succession. Eur J Soil Biol 50:127–136
Ma TL, Chen H, Wang YF, Kang XM, Tian JQ, Zhou XQ, Zhu QA, Peng CH, Liu LF, Hu J, Zhan W, Zhu EX (2015) Effects of enclosure time on the community composition of methanotrophs in the soils of the Inner Mongolia grasslands. J Soil Sediment 16(3):1022–1031
Mori H, Maruyama F, Kato H, Toyoda A, Dozono A, Ohtsubo Y, Nagata Y, Fujiyama A, Tsuda M, Kurokawa K (2014) Design and experimental application of a novel non-degenerate universal primer set that amplifies prokaryotic 16S rRNA genes with a low possibility to amplify eukaryotic rRNA genes. DNA Res 21(2):217–227
Martin M (2011) Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet J 17(1):392–403
Edgar RC, Haas BJ, Clemente JC, Quince C, Knight R (2011) UCHIME improves sensitivity and speed of chimera detection. Bioinformatics 27(16):2194–2200
Haas BJ, Gevers D, Earl AM, Feldgarden M, Ward DV, Giannoukos G, Ciulla D, Tabbaa D, Highlander SK, Sodergren E, Methé B, DeSantis TZ, Petrosino JF, Knight R, Birren BW (2011) Chimeric 16S rRNA sequence formation and detection in Sanger and 454 pyrosequenced PCR amplicons. Genome Res 21(3):494–504
Edgar RC (2013) UPARSE: highly accurate OTU sequences from microbial amplicon reads. Nat Methods 10(10):996–998
Liu JL, Ha VN, Shen Z, Zhu HL, Zhao F, Zhao Z (2018) Response of the rhizosphere microbial community to fine root and soil parameters following Robinia pseudoacacia L. afforestation. Appl Soil Ecol 132:11–19
Cesco S, Mimmo T, Tonon G, Tomasi N, Pinton R, Terzano R, Neumann G, WeisskopfL RG, Landi L, Nannipperi P (2012) Plant-borne flavonoids released into the rhizosphere: impact on soil bio-activities related to plant nutrition: a review. Biol Fertil Soils 48(2):123–149
Li Y, Yang XD, Qin L, Lü GH, He XM, Zhang XN (2018) The bacterial diversity and community structures in rhizosphere soil of two halophytes, Lycium ruthenicum and Kalidium capsicum. Acta Ecol Sin 38(9):3118–3131 (in Chinese)
Kowalchuk GA, Buma DS, Boer WD, Klinkhamer PGL, Veen JAV (2002) Effects of above-ground plant species composition and diversity on the diversity of soil-borne microorganisms. Anton Leeuw Int J G 81(1–4):509–520
Ziegler M, Engel M, Welzl G, Schloter M (2013) Development of a simple root model to study the effects of single exudates on the development of bacterial community structure. J Microbiol Methods 94(1):30–36
HHe YH, Wu ZS, Wang WF, Liu XC, Ye BC (2019) Bacterial community and phosphorus species changes in pepper rhizosphere soils after Pseudomonas putida Rs-198 inoculation. Rhizosphere 11:100164. https://doi.org/10.1016/j.rhisph.2019.100164
Saleem M, Law AD, Sahib MR, Pervaiz ZH, Zhang Q (2018) Impact of root system architecture on rhizosphere and root microbiome. Rhizosphere 6:47–51
Meier IC, Avis PG, Phillips RP (2013) Fungal communities influence root exudation rates in pine seedlings. FEMS Microbiol Ecol 83:585–595
Hmaeid N, Wali M, Metoui-Ben Mahmoud O, Pueyo JJ, Ghnaya T, Abdelly C (2019) Efficient rhizobacteria promote growth and alleviate NaCl-induced stress in the plant species Sulla carnosa. Appl Soil Ecol 133:104–113
Li Y, He XM, Yang XD, Zhang XN, Lü GH (2018) The microbial community diversity of the rhizosphere and bulk soils of Lycium ruthenicum in different habitats. Acta Ecol Sin 38(17):5983–5995 (in Chinese)
Shui Y, Xu ZH, Liu GF (2019) Effect of depth on bacterial diversity in saline-alkali soil in Shizuishan region in Ningxia. Acta Ecol Sin 39(10):3597–3606 (in Chinese)
Liu JL, Ha VN, Shen Z, Zhu HL, Zhao F, Zhao Z (2018) Characteristics of bulk and rhizosphere soil microbial community in an ancient Platycladus orientalis forest. Appl Soil Ecol 132:91–98
Nie M, Zhang XD, Wang JQ, Jiang LF, Yang J, Quan ZX, Cui XH, Fang CM, Li B (2009) Rhizosphere effects on soil bacterial abundance and diversity in the Yellow River Deltaic ecosystem as influenced by petroleum contamination and soil salinization. Soil Biol Biochem 41(12):2535–2542
McNear DH Jr (2013) The rhizosphere-roots, soil and everything in between. Nat Educ Knowl 4:1
Shi L, Du NS, Shu S, Sun J, Li SZ, Guo SR (2017) Paenibacillus polymyxa NSY50 suppresses Fusarium wilt in cucumbers by regulating the rhizospheric microbial community. Sci Rep 7:41234
Albertsen M, Hansen LBS, Saunders AM, Nielsen PH, Nielsen KL (2012) A metagenome of a full-scale microbial community carrying out enhanced biological phosphorus removal. ISME J 6(6):1094–1106
Acknowledgements
We thanks Mrs. Lin Jiang for his help with the production of Fig. S1 by ArcGIS and we would like to thank Editage (www.editage.cn) for English language editing.
Funding
This work was supported by the National Natural Science Foundation of China [Grant Nos. 31660194, 31570595] and Scientific Research Foundation of China West Normal University [Grant Number 18Q045].
Author information
Authors and Affiliations
Contributions
YJ designed research, performed research, analyzed data, and wrote the manuscript; YW participated in the whole experimental process and made important contributions to the experiment; GC contributed to the conception of the study; ZY helped perform the analysis with constructive discussions; MW has made important contributions to the analysis of the manuscript.
Corresponding authors
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Ethical Approval
This article does not contain any studies with human participants or animals performed by any of the authors.
Informed Consent
Informed consent was obtained from all individual participants included in the study.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Jiao, Y., Chu, G., Yang, Z. et al. Bacterial Diversity in the Rhizosphere of Anabasis aphylla in the Gurbantunggut Desert, China. Curr Microbiol 77, 3750–3759 (2020). https://doi.org/10.1007/s00284-020-02177-y
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00284-020-02177-y