Abstract
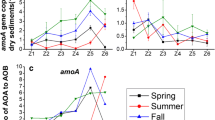
Nitrospira is the most diverse genus of nitrite-oxidizing bacteria, and its members are widely spread in various natural and engineered ecosystems. In this study, the phylogenetic diversity of Nitrospira and monthly changes of its abundance from Zhanjiang Bay were investigated. Phylogenetic analysis showed that among 58 OTUs with high abundance, 74% were not affiliated with any previously described Nitrospira species, revealing a previously unrecognized diversity of coastal Nitrospira. The abundances of both Nitrospira and Nitrospina exhibited a significantly monthly change. During most of the months, abundance of Nitrospina was greater than that of Nitrospira. In particle-attached communities, either abundance of Nitrospina or Nitrospira was highly correlated with that of ammonia-oxidizing archaea (AOA), whereas abundance of ammonia-oxidizing bacteria was only highly correlated with that of Nitrospina. In free-living communities, either abundance of Nitrospina or Nitrospira was correlated only with that of AOA. These results suggest that both Nitrospira and Nitrospina can be involved in nitrite oxidation by coupling with AOA, but Nitrospina may play a greater role than Nitrospira in this tropical bay.




Similar content being viewed by others
Data Availability
All raw sequence reads in this study were deposited in the NCBI Sequence Read Archive (SRA) under accession number PRJNA606648.
References
Pester M, Maixner F, Berry D, Rattei T, Koch H, Lucker S, Nowka B, Richter A, Spieck E, Lebedeva E, Loy A, Wagner M, Daims H (2014) NxrB encoding the beta subunit of nitrite oxidoreductase as functional and phylogenetic marker for nitrite-oxidizing Nitrospira. Environ Microbiol 16:3055–3071
Pajares S, Ramos R (2019) Processes and microorganisms involved in the marine nitrogen cycle: knowledge and gaps. Front Mar Sci 6:e00739
Daims H, Lebedeva EV, Pjevac P, Han P, Herbold C, Albertsen M, Jehmlich N, Palatinszky M, Vierheilig J, Bulaev A, Kirkegaard RH, von Bergen M, Rattei T, Bendinger B, Nielsen PH, Wagner M (2015) Complete nitrification by Nitrospira bacteria. Nature 528:504–509
van Kessel MAHJ, Speth DR, Albertsen M, Nielsen PH, Op den Camp HJM, Kartal B, Jetten MSM, Lucker S (2015) Complete nitrification by a single microorganism. Nature 528:555–559
Parks DH, Rinke C, Chuvochina M, Chaumeil PA, Woodcroft BJ, Evans PN, Hugenholtz P, Tyson GW (2017) Recovery of nearly 8,000 metagenome-assembled genomes substantially expands the tree of life. Nat Microbiol 2:1533–1542
Orellana LH, Chee-Sanford JC, Sanford RA, Loffler FE, Konstantinidis KT (2018) Year-round shotgun metagenomes reveal stable microbial communities in agricultural soils and novel ammonia oxidizers responding to fertilization. Appl Environ Microbiol 84:e01646–e1717
Levipan HA, Molina V, Anguita C, Rain-Franco A, Belmar L, Fernandez C (2016) Variability of nitrifying communities in surface coastal waters of the Eastern South Pacific (~36° S). Environ Microbiol Rep 8:851–864
Wankel SD, Kendall C, Pennington JT, Chavez FP, Paytan A (2007) Nitrification in the euphotic zone as evidenced by nitrate dual isotopic composition: observations from Monterey Bay. California Global Biogeochem Cycle 21:e002723
Yool A, Martin AP, Fernandez C, Clark DR (2007) The significance of nitrification for oceanic new production. Nature 447:999–1002
Beman JM, Popp BN, Alford SE (2012) Quantification of ammonia oxidation rates and ammonia-oxidizing archaea and bacteria at high resolution in the Gulf of California and eastern tropical North Pacific Ocean. Limnol Oceanogr 57:711–726
Pachiadaki MG, Sintes E, Bergauer K, Brown JM, Record NR, Swan BK, Mathyer ME, Hallam SJ, Lopez-Garcia P, Takaki Y, Nunoura T, Woyke T, Herber GJ, Stepanauskas R (2017) Major role of nitrite-oxidizing bacteria in dark ocean carbon fixation. Science 358:1046–1051
Zhang Y, Qin W, Hou L, Zakem EJ, Wan X, Zhao Z, Liu L, Hunt KA, Jiao N, Kao SJ, Tang K, Xie X, Shen J, Li Y, Chen M, Dai X, Liu C, Deng W, Dai M, Ingalls AE, Stahl DA, Herndl GJ (2020) Nitrifier adaptation to low energy flux controls inventory of reduced nitrogen in the dark ocean. Proc Natl Acad Sci USA 117:4823–4830
Prosser JI, Hink L, Gubry-Rangin C, Nicol GW (2020) Nitrous oxide production by ammonia oxidizers: physiological diversity, niche differentiation and potential mitigation strategies. Glob Chang Biol 26:103–118
Breider F, Yoshikawa C, Makabe A, Toyoda S, Wakita M, Matsui Y, Kawagucci S, Fujiki T, Harada N, Yoshida N (2019) Response of N2O production rate to ocean acidification in the western North Pacific. Nat Clim Chang 9:954–958
Daims H, Lucker S, Wagner M (2016) A new perspective on microbes formerly known as nitrite-oxidizing bacteria. Trends Microbiol 24:699–712
Daebeler A, Kitzinger K, Koch H, Herbold CW, Steinfeder M, Schwarz J, Zechmeister T, Karst SM, Albertsen M, Nielsen PH, Wagner M, Daims H (2020) Exploring the upper pH limits of nitrite oxidation: diversity, ecophysiology, and adaptive traits of haloalkalitolerant Nitrospira. ISME J 10:e977850
Kitzinger K, Koch H, Lucker S, Sedlacek CJ, Herbold C, Schwarz J, Daebeler A, Mueller AJ, Lukumbuzya M, Romano S, Leisch N, Karst SM, Kirkegaard R, Albertsen M, Nielsen PH, Wagner M, Daims H (2018) Characterization of the first “candidatus nitrotoga” isolate reveals metabolic versatility and separate evolution of widespread nitrite-oxidizing bacteria. MBio 9:e01186–e11118
Sakoula D, Nowka B, Spieck E, Daims H, Lucker S (2018) The draft genome sequence of "Nitrospira lenta" strain BS10, a nitrite oxidizing bacterium isolated from activated sludge. Stand Genomic Sci 13:32
Spieck E, Bock E (2005) The lithoautotrophic nitrite-oxidizing bcteria. In: George MG (ed) Bergey's manual® of systematic bacteriology, vol 2. Springer, New York, pp 149–153
Alawi M, Lipski A, Sanders T, Eva-Maria-Pfeiffer SE (2007) Cultivation of a novel cold-adapted nitrite oxidizing betaproteobacterium from the Siberian Arctic. ISME J 1:256–264
Sorokin DY, Lucker S, Vejmelkova D, Kostrikina NA, Kleerebezem R, Rijpstra WI, Damste JS, Le Paslier D, Muyzer G, Wagner M, van Loosdrecht MC, Daims H (2012) Nitrification expanded: discovery, physiology and genomics of a nitrite-oxidizing bacterium from the phylum Chloroflexi. ISME J 6:2245–2256
Ngugi DK, Blom J, Stepanauskas R, Stingl U (2016) Diversification and niche adaptations of Nitrospina-like bacteria in the polyextreme interfaces of Red Sea brines. ISME J 10:1383–1399
Hou L, Xie X, Wan X, Kao SJ, Jiao N, Zhang Y (2018) Niche differentiation of ammonia and nitrite oxidizers along a salinity gradient from the Pearl River estuary to the South China Sea. Biogeosciences 15:5169–5187
Poghosyan L, Koch H, Lavy A, Frank J, van Kessel M, Jetten MSM, Banfield JF, Lucker S (2019) Metagenomic recovery of two distinct comammox Nitrospira from the terrestrial subsurface. Environ Microbiol 21:3627–3637
Lebedeva EV, Off S, Zumbragel S, Kruse M, Shagzhina A, Lucker S, Maixner F, Lipski A, Daims H, Spieck E (2011) Isolation and characterization of a moderately thermophilic nitrite-oxidizing bacterium from a geothermal spring. FEMS Microbiol Ecol 75:195–204
Daims H, Nielsen JL, Nielsen PH, Schleifer KH, Wagner M (2001) In situ characterization of Nitrospira-like nitrite-oxidizing bacteria active in wastewater treatment plants. Appl Environ Microbiol 67:5273–5284
Wertz S, Poly F, Le Roux X, Degrange V (2008) Development and application of a PCR-denaturing gradient gel electrophoresis tool to study the diversity of Nitrobacter-like nxrA sequences in soil. FEMS Microbiol Ecol 63:261–271
Helm I, Jalukse L, Vilbaste M, Leito I (2009) Micro-Winkler titration method for dissolved oxygen concentration measurement. Anal Chim Acta 648:167–173
Mantoura RFC, Woodward EMS (1983) Optimization of the indophenol blue method for the automated determination of ammonia in estuarine waters. Estuar Coast Shelf Sci 17:219–224
Hydes DJ, Hill NC (1985) Determination of nitrate in seawater nitrate to nitrite reduction with copper-cadmium alloy. Estuar Coast Shelf Sci 21:127–130
Stewart BM, Elliott PAW (1996) Systematic salt effects in the automated determination of nutrients in seawater. Water Res 30:869–874
Zheng Y, Hou L, Liu M, Lu M, Zhao H, Yin G, Zhou J (2013) Diversity, abundance, and activity of ammonia-oxidizing bacteria and archaea in Chongming eastern intertidal sediments. Appl Microbiol Biotechnol 97:8351–8363
Bolger AM, Lohse M, Usadel B (2014) Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 30:2114–2120
Magoc T, Salzberg SL (2011) FLASH: fast length adjustment of short reads to improve genome assemblies. Bioinformatics 27:2957–2963
Schloss PD, Westcott SL, Ryabin T, Hall JR, Hartmann M, Hollister EB, Lesniewski RA, Oakley BB, Parks DH, Robinson CJ, Sahl JW, Stres B, Thallinger GG, Van Horn DJ, Weber CF (2009) Introducing mothur: open-source, platform-independent, community-supported software for describing and comparing microbial communities. Appl Environ Microbiol 75:7537–7541
Edgar RC, Haas BJ, Clemente JC, Quince C, Knight R (2011) UCHIME improves sensitivity and speed of chimera detection. Bioinformatics 27:2194–2200
Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S (2011) MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol Biol Evol 28:2731–2739
Watson SW, Bock E, Valois FW, Waterbury JB, Schlosser U (1986) Nitrospira marina gen. nov. sp. nov.: a chemolithotrophic nitrite-oxidizing bacterium. Arch Microbiol 144:1–7
Lebedeva EV, Alawi M, Maixner F, Jozsa PG, Daims H, Spieck E (2008) Physiological and phylogenetic characterization of a novel lithoautotrophic nitrite-oxidizing bacterium, 'Candidatus Nitrospira bockiana'. Int J Syst Evol Microbiol 58:242–250
Palatinszky M, Herbold C, Jehmlich N, Pogoda M, Han P, von Bergen M, Lagkouvardos I, Karst SM, Galushko A, Koch H, Berry D, Daims H, Wagner M (2015) Cyanate as an energy source for nitrifiers. Nature 524:105–108
Kitzinger K, Padilla CC, Marchant HK, Hach PF, Herbold CW, Kidane AT, Konneke M, Littmann S, Mooshammer M, Niggemann J, Petrov S, Richter A, Stewart FJ, Wagner M, Kuypers MMM, Bristow LA (2019) Cyanate and urea are substrates for nitrification by Thaumarchaeota in the marine environment. Nat Microbiol 4:234–243
Navada S, Vadstein O, Tveten A-K, Verstege GC, Terjesen BF, Mota VC, Venkataraman V, Gaumet F, Mikkelsen Ø, Kamstra A (2019) Influence of rate of salinity increase on nitrifying biofilms. J Clean Prod 238:117835
Lucker S, Nowka B, Rattei T, Spieck E, Daims H (2013) The genome of Nitrospina gracilis illuminates the metabolism and evolution of the major marine nitrite oxidizer. Front Microbiol 4:27
Mincer TJ, Church MJ, Taylor LT, Preston C, Kar DM, DeLong EF (2007) Quantitative distribution of presumptive archaeal and bacterial nitrifiers in Monterey Bay and the North Pacific Subtropical Gyre. Environ Microbiol 9:1162–1175
Santoro AE, Richter RA, Dupont CL (2019) Planktonic marine archaea. Ann Rev Mar Sci 11:131–158
Ijichi M, Itoh H, Hamasaki K (2019) Vertical distribution of particle-associated and free-living ammonia-oxidizing archaea in Suruga Bay, a deep coastal embayment of Japan. Arch Microbiol 201:1141–1146
Wan XS, Sheng HX, Dai M, Zhang Y, Shi D, Trull TW, Zhu Y, Lomas MW, Kao SJ (2018) Ambient nitrate switches the ammonium consumption pathway in the euphotic ocean. Nat Commun 9:915
Acknowledgements
We thank Qingmei Zhu and Fajin Chen for providing partial nutrient data in water from Zhanjiang Bay.
Funding
This study was funded by the National Natural Science Foundation of China (41971125 and 41725002), the Guangdong Natural Science Foundation (2018A030313164), and the Innovation Study Project of East China Normal University.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no competing interests.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Mao, TQ., Li, YQ., Dong, HP. et al. Spatio-Temporal Variations in the Abundance and Community Structure of Nitrospira in a Tropical Bay. Curr Microbiol 77, 3492–3503 (2020). https://doi.org/10.1007/s00284-020-02193-y
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00284-020-02193-y